

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0055.3
(355 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
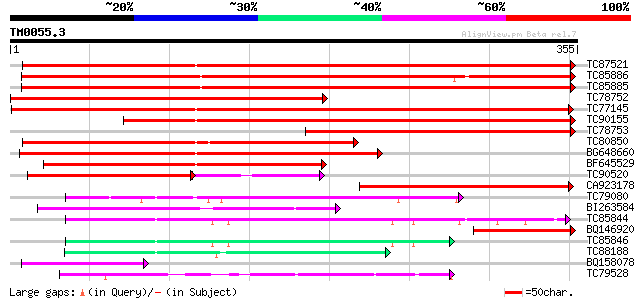
Score E
Sequences producing significant alignments: (bits) Value
TC87521 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydro... 371 e-103
TC85886 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydro... 360 e-100
TC85885 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydro... 358 1e-99
TC78752 similar to GP|22857590|gb|AAN09864.1 putative cinnamyl-a... 344 3e-95
TC77145 homologue to SP|P31656|CADH_MEDSA Cinnamyl-alcohol dehyd... 315 2e-86
TC90155 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydro... 275 1e-74
TC78753 similar to PIR|E96751|E96751 hypothetical protein F28P22... 266 9e-72
TC80850 homologue to SP|O82515|MTD_MEDSA Probable mannitol dehyd... 256 9e-69
BG648660 similar to SP|Q9ZRF1|MTD_ Probable mannitol dehydrogena... 244 4e-65
BF645529 weakly similar to GP|22475166|gb putative sinapyl alcoh... 194 6e-50
TC90520 weakly similar to PIR|T08581|T08581 cinnamyl-alcohol deh... 124 3e-34
CA923178 weakly similar to SP|P93257|MTD_ Probable mannitol dehy... 110 1e-24
TC79080 similar to GP|10177043|dbj|BAB10455. alcohol dehydrogena... 98 4e-21
BI263584 similar to SP|Q9P6C8|ADH1 Alcohol dehydrogenase I (EC 1... 95 4e-20
TC85844 homologue to SP|P12886|ADH1_PEA Alcohol dehydrogenase 1 ... 86 3e-17
BQ146920 similar to GP|8099340|gb|A ELI3 {Lycopersicon esculentu... 75 5e-14
TC85846 homologue to SP|P12886|ADH1_PEA Alcohol dehydrogenase 1 ... 73 2e-13
TC88188 homologue to SP|P80572|ADHX_PEA Alcohol dehydrogenase cl... 72 4e-13
BQ158078 similar to GP|10187159|emb cDNA~Strawberry alcohol dehy... 71 7e-13
TC79528 similar to GP|15808674|gb|AAL06644.1 putative quinone ox... 67 1e-11
>TC87521 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (98%)
Length = 1433
Score = 371 bits (952), Expect = e-103
Identities = 183/347 (52%), Positives = 245/347 (69%), Gaps = 1/347 (0%)
Frame = +2
Query: 9 ECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGNSVYPCVPG 68
+ GWAA DTSGVLSP+ FSRR G+ DV K+ +CG+C++D+ +N G S YP VPG
Sbjct: 101 KAFGWAARDTSGVLSPFNFSRRETGEKDVAFKVLYCGICHSDLHMIKNEWGMSTYPLVPG 280
Query: 69 HEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFTFNGVDHD 128
HEIAGIVT+VGS V F +GD VGVG V+SCR C+ C++ LE++C K T++ D
Sbjct: 281 HEIAGIVTEVGSKVEKFKIGDKVGVGCLVDSCRACQNCEENLENYCPK-QTNTYSAKYSD 457
Query: 129 GTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGVI 188
G+IT GGYS +V E + IP PL SAAPLLCAGITVYSP+ +++PG ++G++
Sbjct: 458 GSITYGGYSDSMVADEHFIVHIPDGLPLESAAPLLCAGITVYSPLRYFGLDKPGMNIGIV 637
Query: 189 GLGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRALAKSLDF 248
GLGGLGH+ VKF KAFG NVTV STS +K++EA+ LGAD F++S DQ++M+A +LD
Sbjct: 638 GLGGLGHLGVKFAKAFGANVTVISTSPNKEKEAIENLGADSFLISHDQDKMQAAMGTLDG 817
Query: 249 IVDTASGPHSFDPYMALLKSFGVLALVGFPGEIKIHPGL-LIMGSRTVSGSGVGGTKEIR 307
I+DT S H P + LLK G L +VG P + P + LIMG +T+SGSG+GG KE +
Sbjct: 818 IIDTVSADHPLLPLVGLLKYHGKLVMVGAPDKPPELPHIPLIMGRKTISGSGIGGMKETQ 997
Query: 308 DMIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDIENSL 354
+MI+F + I P+IEV+P+ Y N A+ R++K DVKYRFV+DI N+L
Sbjct: 998 EMIDFAAKHNIKPDIEVIPVDYVNTAMKRLLKADVKYRFVLDIGNTL 1138
>TC85886 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (94%)
Length = 1395
Score = 360 bits (923), Expect = e-100
Identities = 182/350 (52%), Positives = 241/350 (68%), Gaps = 3/350 (0%)
Frame = +2
Query: 8 EECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGNSVYPCVP 67
++ GWAA D+SGVLSP+ F RR G+ DV K+ +CG+C++D+ +N G + YP VP
Sbjct: 101 KKAFGWAARDSSGVLSPFHFFRRETGEKDVAFKVLYCGICHSDLHIMQNEWGKTTYPLVP 280
Query: 68 GHEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFTFNGVDH 127
GHE+ G+VT+VGS V F VGD VGVG V+SCR CE C D +E++C K + TFNG
Sbjct: 281 GHELTGVVTEVGSKVKKFKVGDKVGVGYMVDSCRSCENCADDIENYCTKYTQ-TFNGKSR 457
Query: 128 DGTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGV 187
DGTIT GG+S +V E + IP S PL A PLLCAG+TVYSP+ +++PG ++GV
Sbjct: 458 DGTITYGGFSDSMVADEHFVIRIPDSLPLDGAGPLLCAGVTVYSPLRHFGLDKPGMNIGV 637
Query: 188 IGLGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRALAKSLD 247
+GLGGLGHMAVKF KAFG VTV STS K++EA+ LGAD F+VS D E+M+A +L+
Sbjct: 638 VGLGGLGHMAVKFAKAFGAKVTVISTSPKKEKEAIEHLGADSFLVSRDPEQMQAATSTLN 817
Query: 248 FIVDTASGPHSFDPYMALLKSFGVLALVGF---PGEIKIHPGLLIMGSRTVSGSGVGGTK 304
I+DT S H P + LLKS G L +VG P E+ I L+ G ++++GS +GG K
Sbjct: 818 GIIDTVSASHPVVPLIGLLKSNGKLVMVGAVAKPLELPIFS--LLGGRKSIAGSLIGGIK 991
Query: 305 EIRDMIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDIENSL 354
E ++MI+F + + P IEVVPI Y N A++R++K DVKYRFVIDI NSL
Sbjct: 992 ETQEMIDFAAKHNVTPEIEVVPIDYVNTAMERLVKGDVKYRFVIDIGNSL 1141
>TC85885 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (97%)
Length = 1330
Score = 358 bits (920), Expect = 1e-99
Identities = 181/348 (52%), Positives = 244/348 (70%), Gaps = 1/348 (0%)
Frame = +3
Query: 8 EECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGNSVYPCVP 67
++ GWAA D+SGVLSP+ FSRR G+ DV K+ +CG+C+ D+ +N GNS+YP VP
Sbjct: 99 KKAFGWAARDSSGVLSPFNFSRRETGEKDVAFKVLYCGICHTDLHMMKNEWGNSIYPLVP 278
Query: 68 GHEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFTFNGVDH 127
GHE+AGIVT+VGS V F VGD VGVG V+SCR CE C + LE++C + +V T
Sbjct: 279 GHELAGIVTEVGSKVEKFKVGDKVGVGYMVDSCRSCENCAEDLENYCPQQTV-TCGAKYR 455
Query: 128 DGTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGV 187
DG++T GGYS +V E + IP S PL A PLLCAG+TVYSP+ ++++PG ++GV
Sbjct: 456 DGSVTYGGYSDSMVADEHFVIRIPDSLPLDVAGPLLCAGVTVYSPLRHFQLDKPGMNIGV 635
Query: 188 IGLGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRALAKSLD 247
+GLGGLGHMAVKF KAFG NVTV STS SK++EA+ LGAD F+VS D ++M+A +L+
Sbjct: 636 VGLGGLGHMAVKFAKAFGANVTVISTSPSKEKEAIEHLGADSFLVSRDPDQMQAAMGTLN 815
Query: 248 FIVDTASGPHSFDPYMALLKSFGVLALVGFPGE-IKIHPGLLIMGSRTVSGSGVGGTKEI 306
I+DT S H P + LLKS G L +VG + +++ L+ G + V+GS +GG KE
Sbjct: 816 GIIDTVSASHPILPLIGLLKSNGKLVMVGGVAKPLELPVFSLLGGRKLVAGSLIGGIKET 995
Query: 307 RDMIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDIENSL 354
++MI+F + + P+IEVVPI Y N A++R+ K DVKYRFVIDI N+L
Sbjct: 996 QEMIDFAAEHNVTPDIEVVPIDYVNTAMERLEKADVKYRFVIDIGNTL 1139
>TC78752 similar to GP|22857590|gb|AAN09864.1 putative cinnamyl-alcohol
dehydrogenase {Oryza sativa (japonica cultivar-group)},
partial (50%)
Length = 668
Score = 344 bits (882), Expect = 3e-95
Identities = 157/199 (78%), Positives = 174/199 (86%)
Frame = +2
Query: 1 MSSEGASEECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGN 60
MSSEG E+CL WAA D SGVLSPYKF+RR +G +DVYVKI+HCGVCYADV W +N G+
Sbjct: 44 MSSEGVGEDCLAWAARDASGVLSPYKFNRRELGSEDVYVKITHCGVCYADVIWAKNKHGD 223
Query: 61 SVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVF 120
S YP VPGHEIAG+V KVG NV F VGDHVGVGTY+NSCR+CE+C+D E HCVKGSV+
Sbjct: 224 SKYPVVPGHEIAGVVAKVGPNVQRFKVGDHVGVGTYINSCRECEYCNDRFEVHCVKGSVY 403
Query: 121 TFNGVDHDGTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQ 180
TFNGVD+DGTITKGGYS IVVHERYCFLIPKS+PLASA PLLCAGITVYSPM+RH MNQ
Sbjct: 404 TFNGVDYDGTITKGGYSTSIVVHERYCFLIPKSHPLASAGPLLCAGITVYSPMIRHNMNQ 583
Query: 181 PGKSLGVIGLGGLGHMAVK 199
PGKSLGV+GLGGLGHMAVK
Sbjct: 584 PGKSLGVVGLGGLGHMAVK 640
>TC77145 homologue to SP|P31656|CADH_MEDSA Cinnamyl-alcohol dehydrogenase
(EC 1.1.1.195) (CAD). [Alfalfa] {Medicago sativa},
complete
Length = 1384
Score = 315 bits (806), Expect = 2e-86
Identities = 160/353 (45%), Positives = 229/353 (64%), Gaps = 1/353 (0%)
Frame = +1
Query: 2 SSEGASEECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGNS 61
S E A +G AA D SG+L+PY ++ R G DDVY+KI +CGVC++D+ +N LG S
Sbjct: 58 SIEVAERTTVGLAAKDPSGILTPYTYTLRNTGPDDVYIKIHYCGVCHSDLHQIKNDLGMS 237
Query: 62 VYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFT 121
YP VPGHE+ G V +VGSNV F VG+ VGVG V C+ C CD +E +C K +++
Sbjct: 238 NYPMVPGHEVVGEVLEVGSNVTRFKVGEIVGVGLLVGCCKSCRACDSEIEQYCNK-KIWS 414
Query: 122 FNGVDHDGTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQP 181
+N V DG IT+GG+++ VV +++ IP+ APLLCAG+TVYSP+ + P
Sbjct: 415 YNDVYTDGKITQGGFAESTVVEQKFVVKIPEGLAPEQVAPLLCAGVTVYSPLSHFGLKTP 594
Query: 182 GKSLGVIGLGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRA 241
G G++GLGG+GHM VK KAFG +VTV S+S KK+EAL LGAD ++VSSD M+
Sbjct: 595 GLRGGILGLGGVGHMGVKVAKAFGHHVTVISSSDKKKKEALEDLGADSYLVSSDTVGMQE 774
Query: 242 LAKSLDFIVDTASGPHSFDPYMALLKSFGVLALVG-FPGEIKIHPGLLIMGSRTVSGSGV 300
A SLD+I+DT H +PY++LLK G L L+G ++ ++++G ++++GS V
Sbjct: 775 AADSLDYIIDTVPVGHPLEPYLSLLKIDGKLILMGVINTPLQFVTPMVMLGRKSITGSFV 954
Query: 301 GGTKEIRDMIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDIENS 353
G KE +M+EF + IE+V + Y N+A +R+ K DV+YRFV+D++ S
Sbjct: 955 GSVKETEEMLEFWKEKGLSSMIEIVTMDYINKAFERLEKNDVRYRFVVDVKGS 1113
>TC90155 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (78%)
Length = 1051
Score = 275 bits (704), Expect = 1e-74
Identities = 144/284 (50%), Positives = 191/284 (66%), Gaps = 1/284 (0%)
Frame = +2
Query: 72 AGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFTFNGVDHDGTI 131
A IVT+VGS V F VGD VGVG ++SCR C+ C+D LE++C K V T DGT+
Sbjct: 2 AAIVTEVGSKVEKFKVGDKVGVGYLIDSCRSCQDCNDNLENYCPK-FVVTCGAKYRDGTV 178
Query: 132 TKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGVIGLG 191
T GGYS +V E + IP + PL A PLLCAG+TVYSP+ +++PG +GV+GLG
Sbjct: 179 TYGGYSDSMVADEHFVIRIPDNIPLEFAGPLLCAGVTVYSPLRFFGLDKPGLHIGVVGLG 358
Query: 192 GLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRALAKSLDFIVD 251
GLGHMAVKF KAFG NVTV STS +K++EA+ LGAD F++SSD ++++ +LD I+D
Sbjct: 359 GLGHMAVKFAKAFGANVTVISTSPNKEKEAIEHLGADSFLISSDPKQIQGAIGTLDGIID 538
Query: 252 TASGPHSFDPYMALLKSFGVLALVG-FPGEIKIHPGLLIMGSRTVSGSGVGGTKEIRDMI 310
T S H P + LLKS G L ++G +++ LI G + ++GS VGG KE ++MI
Sbjct: 539 TVSAVHPLLPMIGLLKSHGKLIMLGVIVQPLQLPEYTLIQGRKILAGSQVGGLKETQEMI 718
Query: 311 EFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDIENSL 354
F + + P+IEVVPI Y N A+ R+ K DVKYRFVIDI N+L
Sbjct: 719 NFAAEHNVKPDIEVVPIDYVNTAMQRLAKGDVKYRFVIDIGNTL 850
>TC78753 similar to PIR|E96751|E96751 hypothetical protein F28P22.13
[imported] - Arabidopsis thaliana, partial (47%)
Length = 865
Score = 266 bits (680), Expect = 9e-72
Identities = 131/169 (77%), Positives = 151/169 (88%)
Frame = +3
Query: 186 GVIGLGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRALAKS 245
GV+GLGGLGHMAVKFGKAFGL VTVFSTS+SKKEEALSLLGAD+FVVSS+QE+MRALAKS
Sbjct: 3 GVVGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSLLGADQFVVSSNQEDMRALAKS 182
Query: 246 LDFIVDTASGPHSFDPYMALLKSFGVLALVGFPGEIKIHPGLLIMGSRTVSGSGVGGTKE 305
LDFI+DTASG H FDPYM+LLK GVL LVGFP E+K P L +GSRTV+GS GGTKE
Sbjct: 183 LDFIIDTASGDHLFDPYMSLLKISGVLVLVGFPSEVKFSPASLNLGSRTVAGSVTGGTKE 362
Query: 306 IRDMIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDIENSL 354
I++M++FC AN IHP+IE++PI Y+NEAL+RV+ KDVKYRFVIDIENSL
Sbjct: 363 IQEMVDFCAANGIHPDIELIPIGYSNEALERVVNKDVKYRFVIDIENSL 509
>TC80850 homologue to SP|O82515|MTD_MEDSA Probable mannitol dehydrogenase
(EC 1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Alfalfa], partial (61%)
Length = 710
Score = 256 bits (654), Expect = 9e-69
Identities = 127/210 (60%), Positives = 153/210 (72%)
Frame = +1
Query: 9 ECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGNSVYPCVPG 68
+ GWAA DTSG LSP+ FSRR GDDDV VKI +CGVC++D+ +N G + YP VPG
Sbjct: 85 KAFGWAARDTSGTLSPFHFSRRENGDDDVSVKILYCGVCHSDLHTLKNDWGFTTYPVVPG 264
Query: 69 HEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFTFNGVDHD 128
HEI G+VTKVG NV F VGD+VGVG V SC+ CE C+ LE +C K VFT+N +
Sbjct: 265 HEIVGVVTKVGINVKKFRVGDNVGVGVIVESCQTCENCNQDLEQYCPK-LVFTYNS-PYK 438
Query: 129 GTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGVI 188
GT T GGYS F+VVH+RY P + PL + APLLCAGITVYSPM + M +PGK LGV
Sbjct: 439 GTRTHGGYSDFVVVHQRYVVQFPDNLPLDAGAPLLCAGITVYSPMKYYGMTEPGKHLGVA 618
Query: 189 GLGGLGHMAVKFGKAFGLNVTVFSTSISKK 218
GLGGLGH+A+KFGKAFGL VTV +TS +K+
Sbjct: 619 GLGGLGHVAIKFGKAFGLKVTVITTSPNKE 708
>BG648660 similar to SP|Q9ZRF1|MTD_ Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (64%)
Length = 721
Score = 244 bits (623), Expect = 4e-65
Identities = 121/227 (53%), Positives = 157/227 (68%)
Frame = +1
Query: 7 SEECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGNSVYPCV 66
S++ GWAA D+SGVLSP+ FSRR + + DV +++ +CG+C++D+ +N G + YP V
Sbjct: 43 SKKAFGWAARDSSGVLSPFNFSRREICEKDVALRVLYCGICHSDLHKAKNEWGTTNYPLV 222
Query: 67 PGHEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFTFNGVD 126
PGHEI GIVT+VGS V F VGD VGVG V+SC C+ C D LE++C + T
Sbjct: 223 PGHEIVGIVTEVGSKVKKFKVGDRVGVGCLVDSCHSCQNCVDNLENYCPQ-LTLTDGSKY 399
Query: 127 HDGTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLG 186
DGT T GG+S +V E + F IP PL +AAPLLCAGITVYSP+ +++P ++G
Sbjct: 400 SDGTSTHGGFSDSMVSDEHFVFRIPDQLPLDAAAPLLCAGITVYSPLRHFGLDKPDMNIG 579
Query: 187 VIGLGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVS 233
V+GLGGLGHMAVKF KAFG NVTV STS K+ EAL L AD F++S
Sbjct: 580 VVGLGGLGHMAVKFAKAFGANVTVISTSPKKENEALEHLRADSFLIS 720
>BF645529 weakly similar to GP|22475166|gb putative sinapyl alcohol
dehydrogenase {Populus tremula x Populus tremuloides},
partial (40%)
Length = 658
Score = 194 bits (492), Expect = 6e-50
Identities = 94/178 (52%), Positives = 124/178 (68%), Gaps = 1/178 (0%)
Frame = +3
Query: 22 LSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGNSVYPCVPGHEIAGIVTKVGSN 81
++PY F RR G DV +KI +CG+C+ DV ++ G ++YP VPGHEI G++TKVGS+
Sbjct: 99 VTPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPVVPGHEITGVITKVGSD 278
Query: 82 VHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFTFNGVDHDGTITKGGYSKFIV 141
V GF GD VGVG SC DCE+C E++C K F +NG+ DG+IT GGYS+ +V
Sbjct: 279 VEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEK-LQFVYNGIFWDGSITYGGYSQMLV 455
Query: 142 VHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKM-NQPGKSLGVIGLGGLGHMAV 198
V RY IP+S P+ +AAPLLCAGITV+SP+ H + + GK +GV+GLG LGHMAV
Sbjct: 456 VDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHXLVSTAGKRIGVVGLGXLGHMAV 629
>TC90520 weakly similar to PIR|T08581|T08581 cinnamyl-alcohol dehydrogenase
(EC 1.1.1.195) CAD1 - Arabidopsis thaliana, partial
(41%)
Length = 618
Score = 124 bits (312), Expect(2) = 3e-34
Identities = 55/105 (52%), Positives = 72/105 (68%)
Frame = +2
Query: 12 GWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGNSVYPCVPGHEI 71
GWAA D+SG ++PY F RR G DV +KI +CG+C+ DV ++ G ++YP VPGHEI
Sbjct: 77 GWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPVVPGHEI 256
Query: 72 AGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVK 116
G++TKVGS+V GF GD VGVG SC DCE+C E++C K
Sbjct: 257 TGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEK 391
Score = 38.5 bits (88), Expect(2) = 3e-34
Identities = 29/84 (34%), Positives = 41/84 (48%), Gaps = 1/84 (1%)
Frame = +3
Query: 115 VKGSVFTFNGVDHDGTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSP-M 173
V+ +NG+ DG+IT GGYS+ +VV R K S+ LC ++ S
Sbjct: 384 VRSCSVVYNGIFWDGSITYGGYSQMLVVDYR------KPTNGCSSTSTLCWNNSIXSL*R 545
Query: 174 MRHKMNQPGKSLGVIGLGGLGHMA 197
++ GK L + LGGLG MA
Sbjct: 546 TTVLVSTAGKKLVXLVLGGLGXMA 617
>CA923178 weakly similar to SP|P93257|MTD_ Probable mannitol dehydrogenase
(EC 1.1.1.255) (NAD-dependent mannitol dehydrogenase).,
partial (34%)
Length = 668
Score = 110 bits (274), Expect = 1e-24
Identities = 57/135 (42%), Positives = 86/135 (63%), Gaps = 1/135 (0%)
Frame = -3
Query: 220 EALSLLGADRFVVSSDQEEMRALAKSLDFIVDTASGPHSFDPYMALLKSFGVLALVGFPG 279
EA LGAD F++S++ ++++A +SLDFI+DT S H+ P + LLK G L +VG P
Sbjct: 663 EAKERLGADDFIISTNPDQLQAAKRSLDFILDTVSADHALLPILELLKVNGTLFIVGAPD 484
Query: 280 EIKIHPGL-LIMGSRTVSGSGVGGTKEIRDMIEFCVANEIHPNIEVVPIQYANEALDRVI 338
+ P LI G R++ G +GG KE ++M++ C + I +IE++ NEA R++
Sbjct: 483 KPLQLPAFPLIFGKRSIKGGIIGGIKETQEMLDVCGKHNITCDIELIKADTINEAFQRLL 304
Query: 339 KKDVKYRFVIDIENS 353
K DV+YRFVIDI N+
Sbjct: 303 KNDVRYRFVIDIANA 259
>TC79080 similar to GP|10177043|dbj|BAB10455. alcohol dehydrogenase-like
protein {Arabidopsis thaliana}, partial (96%)
Length = 1524
Score = 98.2 bits (243), Expect = 4e-21
Identities = 73/281 (25%), Positives = 124/281 (43%), Gaps = 32/281 (11%)
Frame = +3
Query: 36 DVYVKISHCGVCYADVAWTRNTLGNSVYPCVPGHEIAGIVTKVGSN-----VHGFSVGDH 90
++ +K CGVC++D+ + + S PCV GHEI G V + G + + +G
Sbjct: 309 ELLIKTKGCGVCHSDLHVMKGEIPFSS-PCVVGHEITGEVVEHGQHTDSKTIERLPIGSR 485
Query: 91 VGVGTYVNSCRDCEFCDDGLEHHCVKGSVFTFN---GVDHDGTI--------------TK 133
V VG ++ C +C +C G + C + F +N G +DG +
Sbjct: 486 V-VGAFIMPCGNCSYCSKGHDDLCE--AFFAYNRAKGTLYDGETRLFLRGSGNPIYMYSM 656
Query: 134 GGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGVIGLGGL 193
GG +++ VV ++P S P +A L CA T Y M +PG ++ VIG GG+
Sbjct: 657 GGLAEYCVVPANALAVLPNSMPYTESAILGCAVFTAYGAMAHAAEVRPGDTVAVIGTGGV 836
Query: 194 GHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRAL-----AKSLDF 248
G ++ +AFG + + +K E LGA + S+ ++ + + K +D
Sbjct: 837 GSSCLQIARAFGASDIIAVDVQDEKLEKAKTLGATHTINSAKEDPIEKILEITGGKGVDV 1016
Query: 249 IVDTASGPHSFDPYMALLKSFGVLALVGFP-----GEIKIH 284
V+ P +F +K G ++G GE+ I+
Sbjct: 1017AVEALGRPQTFAQCTQSVKDGGKAVMIGLAQAGSLGEVDIN 1139
>BI263584 similar to SP|Q9P6C8|ADH1 Alcohol dehydrogenase I (EC 1.1.1.1).
{Neurospora crassa}, partial (57%)
Length = 682
Score = 95.1 bits (235), Expect = 4e-20
Identities = 57/192 (29%), Positives = 96/192 (49%), Gaps = 2/192 (1%)
Frame = +2
Query: 18 TSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADV-AWTRNTLGNSVYPCVPGHEIAGIVT 76
T+G + + + G D+V V + GVC+ D+ AW + ++ P V GHE AG+V
Sbjct: 113 TAGPIEYKQIPVQKPGPDEVLVNVKFSGVCHTDLHAWQGDWPLDTKLPLVGGHEGAGVVV 292
Query: 77 KVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFTFNGVDHDGTITKGGY 136
G V +G+ VG+ SC C +C + E C + + G G +
Sbjct: 293 ARGDLVKDVKIGEKVGIKWLNGSCLSCSYCQNADESLCAEALL--------SGYTVDGSF 448
Query: 137 SKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGVIGL-GGLGH 195
++ + + IP+ L S +P+LCAGITVY + + + G+S+ ++G GGLG
Sbjct: 449 QQYAIAKAIHVARIPEECDLESISPILCAGITVYKGLKESGV-KAGQSIAIVGAGGGLGS 625
Query: 196 MAVKFGKAFGLN 207
+A ++ KA G++
Sbjct: 626 IAXQYCKAMGIH 661
>TC85844 homologue to SP|P12886|ADH1_PEA Alcohol dehydrogenase 1 (EC
1.1.1.1). [Garden pea] {Pisum sativum}, complete
Length = 1502
Score = 85.5 bits (210), Expect = 3e-17
Identities = 82/346 (23%), Positives = 141/346 (40%), Gaps = 30/346 (8%)
Frame = +2
Query: 36 DVYVKISHCGVCYADVAWTRNTLGNSVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVGT 95
+V +KI +C+ DV + ++P + GHE GIV +G V GDH +
Sbjct: 197 EVRLKILFTSLCHTDVYFWEAKGQTPLFPRIFGHEAGGIVESIGEGVTHLKPGDHA-LPV 373
Query: 96 YVNSCRDCEFCDDGLEHHCVKGSVFTFNGV---DHDGTITKGG-----------YSKFIV 141
+ C DC C + C + T GV D+ + G +S++ V
Sbjct: 374 FTGECGDCPHCKSEESNMCDLLRINTDRGVMINDNQSRFSLKGQPIHHFVGTSTFSEYTV 553
Query: 142 VHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGVIGLGGLGHMAVKFG 201
VH I PL L C T + +PG S+ + GLG +G A +
Sbjct: 554 VHAGCVAKINPDAPLDKVCILSCGICTGLGATINVAKPKPGSSVAIFGLGAVGLAAAEGA 733
Query: 202 KAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEE--MRALAKSLDFIVD-----TAS 254
+ G + + +S + E G + FV D ++ + +A+ + VD T S
Sbjct: 734 RISGASRIIGVDLVSSRFELAKKFGVNEFVNPKDHDKPVQQVIAEMTNGGVDRAVECTGS 913
Query: 255 GPHSFDPYMALLKSFGVLALVGFPGE---IKIHPGLLIMGSRTVSGSGVGGTK---EIRD 308
+ + +GV LVG P + K HP + ++ RT+ G+ G K ++ +
Sbjct: 914 IQAMISAFECVHDGWGVAVLVGVPNKDDAFKTHP-MNLLNERTLKGTFYGNYKPRTDLPN 1090
Query: 309 MIEFCVANEIHPN---IEVVPIQYANEALDRVIKKDVKYRFVIDIE 351
++E + E+ +P N+A D ++K + R +I +E
Sbjct: 1091VVEKYMKGELELEKFITHTIPFSEINKAFDYMLKGE-SIRCIIRME 1225
>BQ146920 similar to GP|8099340|gb|A ELI3 {Lycopersicon esculentum}, partial
(20%)
Length = 686
Score = 74.7 bits (182), Expect = 5e-14
Identities = 33/64 (51%), Positives = 46/64 (71%)
Frame = +2
Query: 291 GSRTVSGSGVGGTKEIRDMIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDI 350
G R GS +GG KE +DM++F + + P IEV+P+ Y N A++R++K DVKYRFVIDI
Sbjct: 8 GER**PGSNIGGMKETQDMLDFAAKHNVKPTIEVIPMDYVNTAMERLLKADVKYRFVIDI 187
Query: 351 ENSL 354
N+L
Sbjct: 188 GNTL 199
>TC85846 homologue to SP|P12886|ADH1_PEA Alcohol dehydrogenase 1 (EC
1.1.1.1). [Garden pea] {Pisum sativum}, partial (78%)
Length = 955
Score = 72.8 bits (177), Expect = 2e-13
Identities = 65/264 (24%), Positives = 103/264 (38%), Gaps = 21/264 (7%)
Frame = +3
Query: 36 DVYVKISHCGVCYADVAWTRNTLGNSVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVGT 95
+V +KI +C+ DV + ++P + GHE GIV VG V GDH +
Sbjct: 153 EVRLKILFTSLCHTDVYFWEAKGQTPLFPRIFGHEAGGIVESVGEGVTHLKPGDHA-LPV 329
Query: 96 YVNSCRDCEFCDDGLEHHCVKGSVFTFNGV---DHDGTITKGG-----------YSKFIV 141
+ C DC C + C + T GV D+ + G +S++ V
Sbjct: 330 FTGECGDCPHCKSEESNMCDLLRINTDRGVMINDNQSRFSIKGQPIHHFVGTSTFSEYTV 509
Query: 142 VHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGVIGLGGLGHMAVKFG 201
VH I PL L C T + +PG S+ + GLG +G A +
Sbjct: 510 VHAGCVAKINPDAPLDKVCILSCGICTGLGATINVAKPKPGSSVAIFGLGAVGLAAAEGA 689
Query: 202 KAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEE--MRALAKSLDFIVD-----TAS 254
+ G + + +S + E G + FV D ++ + +A+ + VD T S
Sbjct: 690 RISGASRIIGVDLVSSRFELAKKFGVNEFVNPKDHDKPVQQVIAEMTNGGVDRAVECTGS 869
Query: 255 GPHSFDPYMALLKSFGVLALVGFP 278
+ + +GV LVG P
Sbjct: 870 IQAMISAFECVHDGWGVAVLVGVP 941
>TC88188 homologue to SP|P80572|ADHX_PEA Alcohol dehydrogenase class III (EC
1.1.1.1) (Glutathione-dependent formaldehyde
dehydrogenase), complete
Length = 1597
Score = 71.6 bits (174), Expect = 4e-13
Identities = 57/221 (25%), Positives = 87/221 (38%), Gaps = 17/221 (7%)
Frame = +1
Query: 35 DDVYVKISHCGVCYADVAWTRNTLGNSVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVG 94
++V ++I +C+ D ++PC+ GHE AGIV VG V GDHV +
Sbjct: 331 NEVRIQILFTALCHTDAYTWSGKDPEGLFPCILGHEAAGIVESVGEGVTDVKPGDHV-IP 507
Query: 95 TYVNSCRDCEFCDDGLEHHCVKGSVFTFNGVDHD-----------------GTITKGGYS 137
Y C +C+FC G + C K T GV + GT T +S
Sbjct: 508 CYQAECGECKFCKSGKTNLCGKVRAATGVGVMMNDRKPRFSIKGKPIYHFMGTST---FS 678
Query: 138 KFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGVIGLGGLGHMA 197
++ VVH+ I L L C T + +PG + + GLG +G
Sbjct: 679 QYTVVHDVSVAKIHPDAALDKVCLLGCGVPTGLGAVWNTAKVEPGSIVAIFGLGTVGLAV 858
Query: 198 VKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEE 238
+ K+ G + + S K + G F+ D E+
Sbjct: 859 AEGAKSAGASRIIGIDIDSNKFDTAKNFGVTEFINPKDHEK 981
>BQ158078 similar to GP|10187159|emb cDNA~Strawberry alcohol dehydrogenase
{Fragaria x ananassa}, partial (19%)
Length = 970
Score = 70.9 bits (172), Expect = 7e-13
Identities = 33/80 (41%), Positives = 48/80 (59%)
Frame = +2
Query: 8 EECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGNSVYPCVP 67
++ GWAA D SGVLSP+ FSRR G+ DV K+ + G+C++D+ +N G YP
Sbjct: 14 KKAFGWAARDPSGVLSPFNFSRRETGEKDVTFKVLY*GICHSDLHMAKNEWGTYFYPLDS 193
Query: 68 GHEIAGIVTKVGSNVHGFSV 87
GHE+A + +V + F V
Sbjct: 194GHELAVLGAEV*NRKFNFQV 253
>TC79528 similar to GP|15808674|gb|AAL06644.1 putative quinone
oxidoreductase {Fragaria x ananassa}, partial (72%)
Length = 834
Score = 67.0 bits (162), Expect = 1e-11
Identities = 71/252 (28%), Positives = 109/252 (43%), Gaps = 5/252 (1%)
Frame = +2
Query: 32 VGDDDVYVKISHCGVCYADVAWTRNTL--GNSVYPCVPGHEIAGIVTKVGSNVHGFSVGD 89
V DD V +K++ + D +S P PG++++G+V KVGS V F VGD
Sbjct: 167 VKDDQVLIKVAAASLNPIDYKRMEGAFKASDSPLPTAPGYDVSGVVVKVGSEVKKFKVGD 346
Query: 90 HVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFTFNGVDHDGTITKGGYSKFIVVHERYCFL 149
V V + LE+ V GS+ + + + ++ H+
Sbjct: 347 EVYGDINVKA----------LEYPKVIGSLAEYTAAE-----------EILLAHK----- 448
Query: 150 IPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGVI-GLGGLGHMAVKFGKAFGLNV 208
PK+ A AA L T Y + R + GKS+ V+ G GG+G ++ K
Sbjct: 449 -PKNLSFAEAASLPLTIETAYEGLERTGFS-AGKSILVLGGAGGVGTHVIQLAKHVYGAS 622
Query: 209 TVFSTSISKKEEALSLLGADRFVVSSDQEEMRALAKSLDFIVDTASGPHSFDPYMALLKS 268
V +TS +KK E LS LGAD + +E L + D + DT + + AL +
Sbjct: 623 KVAATSSTKKLELLSNLGAD-LPIDYTKENFEDLPEKFDVVFDTVG--ETEKAFKALKEG 793
Query: 269 FGVLALV--GFP 278
V+ +V GFP
Sbjct: 794 GKVVTIVPPGFP 829
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.139 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,264,491
Number of Sequences: 36976
Number of extensions: 166716
Number of successful extensions: 745
Number of sequences better than 10.0: 69
Number of HSP's better than 10.0 without gapping: 717
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 720
length of query: 355
length of database: 9,014,727
effective HSP length: 97
effective length of query: 258
effective length of database: 5,428,055
effective search space: 1400438190
effective search space used: 1400438190
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0055.3


