

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
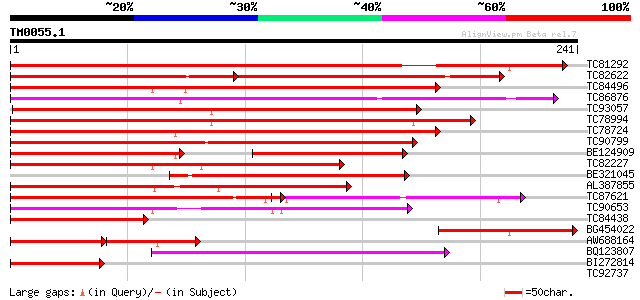
Query= TM0055.1
(241 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC81292 homologue to PIR|T09700|T09700 MADS-box protein - alfalf... 279 8e-76
TC82622 similar to GP|21739238|emb|CAD40993. OSJNBa0072F16.17 {O... 148 7e-70
TC84496 similar to GP|1483232|emb|CAA67969.1 MADS5 protein {Betu... 162 8e-41
TC86876 similar to GP|16973296|emb|CAC80857. C-type MADS box pro... 159 7e-40
TC93057 homologue to GP|23194453|gb|AAN15183.1 MADS box protein ... 153 7e-38
TC78994 similar to SP|O64645|SOC1_ARATH SUPPRESSOR OF CONSTANS O... 148 2e-36
TC78724 similar to GP|4322475|gb|AAD16052.1| putative MADS box t... 148 2e-36
TC90799 similar to GP|7672991|gb|AAF66690.1| MADS-box transcript... 143 7e-35
BE124909 similar to GP|4322475|gb| putative MADS box transcripti... 102 6e-34
TC82227 similar to GP|1483232|emb|CAA67969.1 MADS5 protein {Betu... 140 6e-34
BE321045 similar to PIR|T09700|T097 MADS-box protein - alfalfa (... 137 5e-33
AL387855 similar to GP|1483230|emb MADS4 protein {Betula pendula... 137 5e-33
TC87621 similar to SP|Q9FVC1|SVP_ARATH SHORT VEGETATIVE PHASE pr... 113 1e-31
TC90653 similar to GP|10835358|gb|AAC13695.2 PTD protein {Populu... 108 2e-24
TC84438 homologue to GP|1483232|emb|CAA67969.1 MADS5 protein {Be... 91 5e-19
BG454022 86 2e-17
AW688164 similar to GP|3114588|gb|A MADS box protein {Eucalyptus... 62 5e-16
BQ123807 similar to GP|13810202|em MADS2 protein {Cucumis sativu... 76 1e-14
BI272814 PIR|S71757|S71 MADS box protein DEFH200 - garden snapdr... 64 4e-11
TC92737 weakly similar to GP|15022157|gb|AAK77938.1 MADS box pro... 39 0.001
>TC81292 homologue to PIR|T09700|T09700 MADS-box protein - alfalfa
(fragment), partial (91%)
Length = 800
Score = 279 bits (713), Expect = 8e-76
Identities = 150/238 (63%), Positives = 179/238 (75%), Gaps = 1/238 (0%)
Frame = +2
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
MGRGKI IRRIDNSTSRQVTFSKRRNGLLKKAKEL+ILCDAEVG+++FSST KLY++AST
Sbjct: 71 MGRGKIQIRRIDNSTSRQVTFSKRRNGLLKKAKELAILCDAEVGVMIFSSTAKLYDFAST 250
Query: 61 SMKSVIERYNKLKEEHNQLMNPASELKFWQTEAASLRQQLQYLQECHRQLMGEGLSGLGV 120
SM+SVI+RYNK KEEHNQL + SE+KFWQ EAA LRQQL LQE HRQ+MGE LSGL V
Sbjct: 251 SMRSVIDRYNKTKEEHNQLGSSTSEIKFWQREAAMLRQQLHNLQESHRQIMGEELSGLTV 430
Query: 121 KELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKMELIQKENAEL 180
KELQ LENQLE+SL+GVRMKK+Q+ DEI+EL++KG++IHQEN ELY
Sbjct: 431 KELQGLENQLEISLRGVRMKKEQLFMDEIQELNRKGDIIHQENVELY------------- 571
Query: 181 QKKVYEARSTNEKDAASSPSCTIRNGYDLQA-ISLQLSQPQPQFSEPPAKAVKLGYSL 237
+KVY + N + S + + G D ++LQLSQPQ Q + P+ KLG L
Sbjct: 572 -RKVYGTKDKNGTNRVLSLTNGVGIGDDSNVPVNLQLSQPQQQHYKAPSGTTKLGLQL 742
>TC82622 similar to GP|21739238|emb|CAD40993. OSJNBa0072F16.17 {Oryza
sativa}, partial (78%)
Length = 847
Score = 148 bits (374), Expect(2) = 7e-70
Identities = 74/97 (76%), Positives = 84/97 (86%)
Frame = +3
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
MGRGKI IRRIDN TSRQVTFSKRR GL+KKAKEL+ILCDA+VGL++FSSTGKLYEYA+T
Sbjct: 138 MGRGKIVIRRIDNCTSRQVTFSKRRKGLIKKAKELAILCDAQVGLVIFSSTGKLYEYANT 317
Query: 61 SMKSVIERYNKLKEEHNQLMNPASELKFWQTEAASLR 97
SMKSVIERYN KE+ Q+ NP SE+KFWQ EA +
Sbjct: 318 SMKSVIERYNICKED-QQVTNPESEVKFWQREAVHFK 425
Score = 132 bits (333), Expect(2) = 7e-70
Identities = 67/115 (58%), Positives = 93/115 (80%)
Frame = +1
Query: 96 LRQQLQYLQECHRQLMGEGLSGLGVKELQNLENQLEMSLKGVRMKKDQILTDEIKELHQK 155
LRQQLQ LQE HRQLMGE L GL ++ LQ+LE+QLE+SL+GVRMKK++ILTDEI+EL++K
Sbjct: 421 LRQQLQSLQENHRQLMGEQLYGLSIRNLQDLESQLELSLQGVRMKKEKILTDEIQELNRK 600
Query: 156 GNLIHQENAELYKKMELIQKENAELQKKVYEARSTNEKDAASSPSCTIRNGYDLQ 210
G++IHQEN ELYKK+ L+Q+EN +L KKVY +T+ + A+S + ++ Y ++
Sbjct: 601 GSIIHQENVELYKKVNLLQQENTQLHKKVY--GTTDNEATATSKNAFVQFPYSVR 759
>TC84496 similar to GP|1483232|emb|CAA67969.1 MADS5 protein {Betula
pendula}, partial (74%)
Length = 681
Score = 162 bits (411), Expect = 8e-41
Identities = 84/185 (45%), Positives = 129/185 (69%), Gaps = 2/185 (1%)
Frame = +2
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAS- 59
MGRG++ +++I+N +RQVTFSKRR+GLLKKA E+S+LCDAEV LIVFS+ GKLYEY+S
Sbjct: 26 MGRGRVQLKKIENKINRQVTFSKRRSGLLKKAHEISVLCDAEVALIVFSNKGKLYEYSSD 205
Query: 60 TSMKSVIERYNKLK-EEHNQLMNPASELKFWQTEAASLRQQLQYLQECHRQLMGEGLSGL 118
M+ ++ERY + E + N + + W E A L+ +L+ +Q+ R MGE L GL
Sbjct: 206 PCMEKILERYERYSYAERQHVANDQPQNENWIIEHARLKTRLEVIQKNQRNFMGEELDGL 385
Query: 119 GVKELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKMELIQKENA 178
+KELQ+LE+QL+ +LK +R +K+Q++ + I EL +K + ++N L KM+ +K A
Sbjct: 386 SMKELQHLEHQLDSALKQIRSRKNQLMYESISELSKKDKALQEKNKLLTTKMKEKEKALA 565
Query: 179 ELQKK 183
+L+++
Sbjct: 566 QLEQQ 580
>TC86876 similar to GP|16973296|emb|CAC80857. C-type MADS box protein {Malus
x domestica}, partial (85%)
Length = 2050
Score = 159 bits (403), Expect = 7e-40
Identities = 93/234 (39%), Positives = 141/234 (59%), Gaps = 1/234 (0%)
Frame = +2
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
MGRGKI I+RI+N+T+RQVTF KRRNGLLKKA ELS+LCDAEV L+VFS+ G+LYEYA+
Sbjct: 89 MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALVVFSTRGRLYEYANN 268
Query: 61 SMKSVIERYNK-LKEEHNQLMNPASELKFWQTEAASLRQQLQYLQECHRQLMGEGLSGLG 119
S+++ IERY K N + +F+Q E++ LR+Q++ +Q +R ++GE L L
Sbjct: 269 SVRATIERYKKACAASTNAESVSEANTQFYQQESSKLRRQIRDIQNLNRHILGEALGSLS 448
Query: 120 VKELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKMELIQKENAE 179
+KEL+NLE +LE L VR +K + L +++ + ++ I +N Y + ++ + E A+
Sbjct: 449 LKELKNLEGRLEKGLSRVRSRKHETLFADVEFMQKRE--IELQNHNNYLRAKIAEHERAQ 622
Query: 180 LQKKVYEARSTNEKDAASSPSCTIRNGYDLQAISLQLSQPQPQFSEPPAKAVKL 233
Q+ T + S RN + + L Q Q+S A++L
Sbjct: 623 QQQHNLMPDQTMCDQSLPSSQAYDRNFFPVNL----LGSDQQQYSRQDQTALQL 772
>TC93057 homologue to GP|23194453|gb|AAN15183.1 MADS box protein GHMADS-2
{Gossypium hirsutum}, partial (95%)
Length = 858
Score = 153 bits (386), Expect = 7e-38
Identities = 81/175 (46%), Positives = 118/175 (67%), Gaps = 1/175 (0%)
Frame = +3
Query: 2 GRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYASTS 61
G ++ + I+N+T+RQVTF KRRNGLLKKA ELS+LCDAEV LIVFSS G+LYEY++ +
Sbjct: 33 GGERLR*KGIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYSNNN 212
Query: 62 MKSVIERYNKLKEEHNQLMNPAS-ELKFWQTEAASLRQQLQYLQECHRQLMGEGLSGLGV 120
++S I+RY K +H+ +++Q E+A LRQQ+Q LQ +R LMG+ LS L V
Sbjct: 213 IRSTIDRYKKACSDHSSTTTTTEINAQYYQQESAKLRQQIQMLQNSNRHLMGDALSTLTV 392
Query: 121 KELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKMELIQK 175
KEL+ LEN+LE + +R KK ++L EI+ ++ + EN L K+ +++
Sbjct: 393 KELKQLENRLERGITRIRSKKHEMLLAEIEYFQKREIELENENLCLRTKINDVER 557
>TC78994 similar to SP|O64645|SOC1_ARATH SUPPRESSOR OF CONSTANS
OVEREXPRESSION 1 protein (Agamous-like MADS box protein
AGL20)., partial (83%)
Length = 946
Score = 148 bits (374), Expect = 2e-36
Identities = 84/202 (41%), Positives = 125/202 (61%), Gaps = 4/202 (1%)
Frame = +3
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
M RGK ++RI+N+TSRQVTFSKRRNGLLKKA ELS+LCDAEV LIVFS G+LYE+AS+
Sbjct: 54 MVRGKTQMKRIENATSRQVTFSKRRNGLLKKAFELSVLCDAEVALIVFSPRGRLYEFASS 233
Query: 61 SMKSVIERYNKLKEEHNQLMNPAS--ELKFWQTEAASLRQQLQYLQECHRQLMGEGLSGL 118
S+ IERY +N S + + EA ++ +++ L+ R+L+GEGL
Sbjct: 234 SILETIERYRSHTRINNTPTTSESVENTQQLKEEAENMMKKIDLLETSKRKLLGEGLGSC 413
Query: 119 GVKELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKM--ELIQKE 176
+ ELQ +E QLE S+ +R+KK ++ ++I +L +K + EN L +K Q+
Sbjct: 414 SIDELQKIEQQLEKSINKIRVKKTKVFREQIDQLKEKEKALVAENVRLSEKYGNYSTQES 593
Query: 177 NAELQKKVYEARSTNEKDAASS 198
+ ++ + EA ++ + SS
Sbjct: 594 TKDQRENIAEAEPYADQSSPSS 659
>TC78724 similar to GP|4322475|gb|AAD16052.1| putative MADS box
transcription factor ETL {Eucalyptus globulus subsp.
globulus}, partial (71%)
Length = 1089
Score = 148 bits (373), Expect = 2e-36
Identities = 82/184 (44%), Positives = 120/184 (64%), Gaps = 1/184 (0%)
Frame = +1
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
M RGK ++RI+N ++RQVTFSKRRNGLLKKA ELS+LCDAEV LIVFS+TGKLYE++S+
Sbjct: 226 MVRGKTQMKRIENESNRQVTFSKRRNGLLKKAFELSVLCDAEVALIVFSTTGKLYEFSSS 405
Query: 61 SMKSVIERY-NKLKEEHNQLMNPASELKFWQTEAASLRQQLQYLQECHRQLMGEGLSGLG 119
S+ +ERY K+KE + + ++L++L+ R+L+GE L
Sbjct: 406 SISKTVERYQGKVKELVLSTKGIQENTQHLKECDIDTTKKLEHLELSKRKLLGEELGSCA 585
Query: 120 VKELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKMELIQKENAE 179
ELQ +ENQLE SL +R +K+Q+L ++I++L K L+ +EN L K+ + Q +
Sbjct: 586 FDELQQIENQLERSLSKIRARKNQLLKEQIEKLKDKERLLLEENKRLCKQCGIGQNDCLN 765
Query: 180 LQKK 183
Q++
Sbjct: 766 KQQE 777
>TC90799 similar to GP|7672991|gb|AAF66690.1| MADS-box transcription factor
{Canavalia lineata}, partial (76%)
Length = 653
Score = 143 bits (360), Expect = 7e-35
Identities = 78/173 (45%), Positives = 112/173 (64%)
Frame = +2
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
M R KI I++IDN+T+RQVTFSKRR G+ KKA+ELSILCDAEVGL++FS+TGKLYEYAS+
Sbjct: 71 MARQKIKIKKIDNATARQVTFSKRRRGIFKKAEELSILCDAEVGLVIFSTTGKLYEYASS 250
Query: 61 SMKSVIERYNKLKEEHNQLMNPASELKFWQTEAASLRQQLQYLQECHRQLMGEGLSGLGV 120
+MK +I RY + +L P +++ + A L +++ + R + E GL +
Sbjct: 251 NMKDIITRYGQQSHHITKLDKPL-QVQVEKNMPAELNKEVADRTQQLRGMKSEDFEGLNL 427
Query: 121 KELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKMELI 173
+ LQ LE LE LK V K++ + +EIK L K ++ EN L +KM ++
Sbjct: 428 EGLQQLEKSLESXLKRVIEMKEKKILNEIKALRMKEIMLEXENKHLKQKMAML 586
>BE124909 similar to GP|4322475|gb| putative MADS box transcription factor
ETL {Eucalyptus globulus subsp. globulus}, partial (60%)
Length = 608
Score = 102 bits (254), Expect(2) = 6e-34
Identities = 51/75 (68%), Positives = 64/75 (85%), Gaps = 1/75 (1%)
Frame = +1
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
M RGK ++RI+N ++RQVTFSKRRNGLLKKA ELS+LCDAEV LIVFS+TGKLYE++S+
Sbjct: 148 MVRGKTQMKRIENESNRQVTFSKRRNGLLKKAFELSVLCDAEVALIVFSTTGKLYEFSSS 327
Query: 61 SMKSVIERY-NKLKE 74
S+ +ERY K+KE
Sbjct: 328 SISKTVERYQGKVKE 372
Score = 58.9 bits (141), Expect(2) = 6e-34
Identities = 28/66 (42%), Positives = 44/66 (66%)
Frame = +2
Query: 104 QECHRQLMGEGLSGLGVKELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQEN 163
++ HR+L+GE L ELQ +ENQLE SL +R +K+Q+L ++I++L K L+ +EN
Sbjct: 398 KKIHRKLLGEELGSCAFDELQQIENQLERSLSKIRARKNQLLKEQIEKLKDKERLLLEEN 577
Query: 164 AELYKK 169
L K+
Sbjct: 578 KRLCKQ 595
>TC82227 similar to GP|1483232|emb|CAA67969.1 MADS5 protein {Betula
pendula}, partial (68%)
Length = 681
Score = 140 bits (352), Expect = 6e-34
Identities = 71/144 (49%), Positives = 104/144 (71%), Gaps = 2/144 (1%)
Frame = +1
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAS- 59
MGRG++ ++RI+N +RQVTFSKRR+GLLKKA+E+S+LCDAEV LI+FS+ GKL+EY+S
Sbjct: 130 MGRGRVQLKRIENKINRQVTFSKRRSGLLKKAQEISVLCDAEVALIIFSTKGKLFEYSSD 309
Query: 60 TSMKSVIERYNKLKEEHNQLM-NPASELKFWQTEAASLRQQLQYLQECHRQLMGEGLSGL 118
M+ ++ERY + QL+ + S + W E A L+ +++ LQ R MGE L GL
Sbjct: 310 PCMEKILERYERCSYMERQLVTSEQSPNENWVLEHAKLKARMEVLQRNQRNFMGEDLDGL 489
Query: 119 GVKELQNLENQLEMSLKGVRMKKD 142
G+KELQ+LE QL+ +LK + + ++
Sbjct: 490 GLKELQSLEQQLDSALKQITITEE 561
>BE321045 similar to PIR|T09700|T097 MADS-box protein - alfalfa (fragment),
partial (38%)
Length = 423
Score = 137 bits (344), Expect = 5e-33
Identities = 69/102 (67%), Positives = 86/102 (83%)
Frame = +1
Query: 69 YNKLKEEHNQLMNPASELKFWQTEAASLRQQLQYLQECHRQLMGEGLSGLGVKELQNLEN 128
YN KE+ Q+ NP SE+KFWQ EA LRQQLQ LQE HRQLMGE L GL ++ LQ+LE+
Sbjct: 1 YNICKEDQ-QVTNPESEVKFWQREADILRQQLQSLQENHRQLMGEQLYGLSIRNLQDLES 177
Query: 129 QLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKM 170
QLE+SL+GVRMKK++ILTDEI+EL++KG++IHQEN ELYKK+
Sbjct: 178 QLELSLQGVRMKKEKILTDEIQELNRKGSIIHQENVELYKKV 303
>AL387855 similar to GP|1483230|emb MADS4 protein {Betula pendula}, partial
(51%)
Length = 493
Score = 137 bits (344), Expect = 5e-33
Identities = 71/147 (48%), Positives = 103/147 (69%), Gaps = 2/147 (1%)
Frame = +2
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
MGRG++ ++RI+N TS+QVTF KRR GLLKKA E+S+LCDA+V LI+FS+ GKL+EY+S
Sbjct: 53 MGRGRVQLKRIENKTSQQVTFFKRRTGLLKKANEISVLCDAQVALIMFSTKGKLFEYSSA 232
Query: 61 -SMKSVIERYNKLKEEHNQLMNPASELK-FWQTEAASLRQQLQYLQECHRQLMGEGLSGL 118
SM+ ++ERY ++ H +L +E + W E L ++Q L+ R +G L L
Sbjct: 233 PSMEDILERYE--RQNHTELTGATNETQGNWSFEYMKLTAKVQVLERNLRNFVGNDLDPL 406
Query: 119 GVKELQNLENQLEMSLKGVRMKKDQIL 145
VKELQ+LE QL+ SLK +R +K+Q++
Sbjct: 407 SVKELQSLEQQLDTSLKRIRTRKNQVM 487
>TC87621 similar to SP|Q9FVC1|SVP_ARATH SHORT VEGETATIVE PHASE protein.
[Mouse-ear cress] {Arabidopsis thaliana}, partial (65%)
Length = 1322
Score = 113 bits (283), Expect(2) = 1e-31
Identities = 60/119 (50%), Positives = 85/119 (71%), Gaps = 2/119 (1%)
Frame = +2
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAST 60
M R KI I++I+NST+RQVTFSKRR GL+KKA+ELS+LCDA+V LI+FSSTGKL+EY++
Sbjct: 437 MAREKIQIKKIENSTARQVTFSKRRRGLIKKAEELSVLCDADVALIIFSSTGKLFEYSNL 616
Query: 61 SMKSVIERYNKLKEEHNQLMNPASELKFWQTEAASLRQQLQYLQECH--RQLMGEGLSG 117
SM+ ++ER++ + +L P+ EL+ + S R + Q+ H RQ+ GE L G
Sbjct: 617 SMREILERHHLHSKNLAKLEEPSLELQLVENSNCS-RLSKEVAQKSHQLRQMRGEDLQG 790
Score = 39.7 bits (91), Expect(2) = 1e-31
Identities = 35/115 (30%), Positives = 51/115 (43%), Gaps = 7/115 (6%)
Frame = +3
Query: 112 GEGLS-GLGVKELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKM 170
GE +S GL ++ELQ LE LE+ L V K + + EI EL KG + +EN L K
Sbjct: 771 GERISKGLSLEELQQLEKSLEIGLGRVIETKGEKIMMEINELQTKGRQLMEENNRL--KR 944
Query: 171 ELIQKENAELQKKVYEARSTNEKDAASSPSCTIRNG------YDLQAISLQLSQP 219
+ N ++ V E+ +S + N Y+ SL+L P
Sbjct: 945 HVSGMFNGKMFGGVESENMVTEEGQSSESVTNVYNSTGPPQDYESSDTSLKLGLP 1109
>TC90653 similar to GP|10835358|gb|AAC13695.2 PTD protein {Populus
balsamifera subsp. trichocarpa}, partial (59%)
Length = 845
Score = 108 bits (270), Expect = 2e-24
Identities = 63/183 (34%), Positives = 103/183 (55%), Gaps = 12/183 (6%)
Frame = +2
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAS- 59
MGRGKI I+ I+N T+RQVT+SKRRNG+ KKA ELS+LCDA+V LI+FS K++EY +
Sbjct: 44 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 223
Query: 60 -TSMKSVIERYNKLKEEHNQLMNPASELKFWQTEAASLRQQLQYLQECHRQL-------M 111
S K +I++Y K ++ W++ + + L+ L++ + +L +
Sbjct: 224 GLSTKKIIDQYQK----------TLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRI 373
Query: 112 GEG---LSGLGVKELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYK 168
GEG L L ++L++LE + S+ +R +K ++ +K + Q N L
Sbjct: 374 GEGGMELDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLL 553
Query: 169 KME 171
++E
Sbjct: 554 ELE 562
>TC84438 homologue to GP|1483232|emb|CAA67969.1 MADS5 protein {Betula
pendula}, partial (24%)
Length = 702
Score = 90.5 bits (223), Expect = 5e-19
Identities = 40/59 (67%), Positives = 54/59 (90%)
Frame = +1
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDAEVGLIVFSSTGKLYEYAS 59
MGRG++ ++RI+N +RQVTFSKRR+GLLKKA+E+S+LCDAEV LI+FS+ GKL+EY+S
Sbjct: 223 MGRGRVQLKRIENKINRQVTFSKRRSGLLKKAQEISVLCDAEVALIIFSTKGKLFEYSS 399
>BG454022
Length = 660
Score = 85.5 bits (210), Expect = 2e-17
Identities = 44/60 (73%), Positives = 54/60 (89%), Gaps = 1/60 (1%)
Frame = -3
Query: 183 KVYEARSTNEKDAASSPSCTIRNGYDLQA-ISLQLSQPQPQFSEPPAKAVKLGYSLKLIL 241
+VYEARSTNE++AA++ S TIRNG+DL A ISLQLSQPQPQ+SEP AK +KLGYSL+LI+
Sbjct: 502 QVYEARSTNEENAATNLSRTIRNGFDLHAPISLQLSQPQPQYSEPAAKVMKLGYSLELIM 323
>AW688164 similar to GP|3114588|gb|A MADS box protein {Eucalyptus grandis},
partial (31%)
Length = 549
Score = 62.4 bits (150), Expect(2) = 5e-16
Identities = 28/41 (68%), Positives = 35/41 (85%)
Frame = +1
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCDA 41
MGRG++ + RI+N +RQVTFSKRR+GLLKKA EL +LCDA
Sbjct: 7 MGRGRVVLERIENKINRQVTFSKRRSGLLKKAFELCVLCDA 129
Score = 38.5 bits (88), Expect(2) = 5e-16
Identities = 19/41 (46%), Positives = 28/41 (67%), Gaps = 1/41 (2%)
Frame = +2
Query: 42 EVGLIVFSSTGKLYEYASTS-MKSVIERYNKLKEEHNQLMN 81
EV LI+FSS GKL++Y+ST+ + +IERY + + Q N
Sbjct: 131 EVALIIFSSRGKLFQYSSTTDINKIIERYRQCRYSKPQAGN 253
>BQ123807 similar to GP|13810202|em MADS2 protein {Cucumis sativus}, partial
(88%)
Length = 676
Score = 76.3 bits (186), Expect = 1e-14
Identities = 43/127 (33%), Positives = 67/127 (51%)
Frame = +3
Query: 61 SMKSVIERYNKLKEEHNQLMNPASELKFWQTEAASLRQQLQYLQECHRQLMGEGLSGLGV 120
SM + RY K ++ PA EL+ E L+ + + LQ R L+GE L LG
Sbjct: 6 SMLKTLVRYQKCSYGAVEVNKPAKELESSYREYLKLKARFESLQRTQRNLLGEDLGPLGT 185
Query: 121 KELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAELYKKMELIQKENAEL 180
K+L+ LE QL+ SLK VR K Q + D++ +L K +++ + N L K+E I +
Sbjct: 186 KDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKEHMLVEANRSLSMKLEEININSRNQ 365
Query: 181 QKKVYEA 187
++ +EA
Sbjct: 366 YRQTWEA 386
>BI272814 PIR|S71757|S71 MADS box protein DEFH200 - garden snapdragon,
partial (16%)
Length = 320
Score = 64.3 bits (155), Expect = 4e-11
Identities = 28/40 (70%), Positives = 36/40 (90%)
Frame = +1
Query: 1 MGRGKIAIRRIDNSTSRQVTFSKRRNGLLKKAKELSILCD 40
MGRG++ ++RI+N +RQVTF+KRRNGLLKKA ELS+LCD
Sbjct: 199 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCD 318
>TC92737 weakly similar to GP|15022157|gb|AAK77938.1 MADS box protein-like
protein NGL9 {Medicago sativa}, partial (33%)
Length = 541
Score = 39.3 bits (90), Expect = 0.001
Identities = 23/59 (38%), Positives = 31/59 (51%)
Frame = +3
Query: 108 RQLMGEGLSGLGVKELQNLENQLEMSLKGVRMKKDQILTDEIKELHQKGNLIHQENAEL 166
R L GE ++ L KEL LE LE L GVR KK ++ + + G ++ EN EL
Sbjct: 63 RHLKGEDITSLNYKELMALEESLENGLTGVRDKKMEV----HRMFKRNGKILEDENKEL 227
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.312 0.129 0.346
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,798,399
Number of Sequences: 36976
Number of extensions: 65024
Number of successful extensions: 341
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 330
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 332
length of query: 241
length of database: 9,014,727
effective HSP length: 93
effective length of query: 148
effective length of database: 5,575,959
effective search space: 825241932
effective search space used: 825241932
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0055.1


