

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
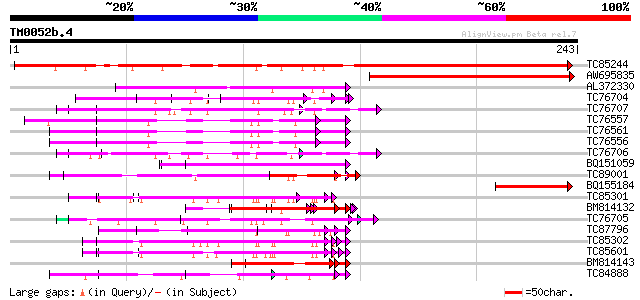
Query= TM0052b.4
(243 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85244 similar to GP|2677824|gb|AAB97140.1| abscisic stress rip... 279 6e-76
AW695835 similar to GP|14582465|gb| putative transcription facto... 151 3e-37
AL372330 similar to GP|21322752|dbj cold shock protein-1 {Tritic... 81 3e-16
TC76704 homologue to SP|Q09134|GRPA_MEDFA Abscisic acid and envi... 80 5e-16
TC76707 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulat... 77 6e-15
TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 73 1e-13
TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 72 2e-13
TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 72 2e-13
TC76706 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulat... 71 3e-13
BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melano... 69 1e-12
TC89001 similar to GP|5726567|gb|AAD48471.1| glycine-rich RNA-bi... 69 2e-12
BQ155184 homologue to GP|11994861|dbj putative ripening protein ... 67 5e-12
TC85301 similar to GP|8096255|dbj|BAA96106.1 TrPRP2 {Trifolium r... 65 2e-11
BM814132 weakly similar to OMNI|MT3615.1 PE_PGRS family protein ... 65 2e-11
TC76705 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulat... 65 3e-11
TC87796 weakly similar to SP|P03211|EBN1_EBV EBNA-1 nuclear prot... 64 4e-11
TC85302 similar to GP|437310|gb|AAA62850.1|| nodulin {Medicago t... 62 2e-10
TC85601 nodulin 61 3e-10
BM814143 GP|21322711|e pherophorin-dz1 protein {Volvox carteri f... 61 3e-10
TC84888 similar to GP|21322752|dbj|BAB78536. cold shock protein-... 60 6e-10
>TC85244 similar to GP|2677824|gb|AAB97140.1| abscisic stress ripening
protein homolog {Prunus armeniaca}, partial (55%)
Length = 1270
Score = 279 bits (714), Expect = 6e-76
Identities = 164/271 (60%), Positives = 188/271 (68%), Gaps = 32/271 (11%)
Frame = +2
Query: 3 EENRDRGLFHHHKNED---RPTDTETGYNKTS-YSTDEPSGGDYDSGYKKSSY--ETSGG 56
EENR RGLFHH K++D RP DT++GYNKTS YST + DY+ KK+SY + SGG
Sbjct: 230 EENRSRGLFHHKKHDDDDNRPIDTDSGYNKTSSYSTGDD---DYN---KKTSYGNDNSGG 391
Query: 57 GYETGYNK-TSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGA----GGGYSTG 111
YETGYNK T +YS +E + GGY SG KT +T GGGYG GGGY G
Sbjct: 392 DYETGYNKNTDNYSNNETS---GGYGSGGAKT--TTGGGYGGGYGDSDTKTTTGGGY--G 550
Query: 112 GGYG-------TGGGGGGEY-------STGGG-GGGY------STGGGGYGNDDSGNRRD 150
GGYG TGGG GG Y +TGGG GGGY +T GGGYG+D RR+
Sbjct: 551 GGYGDSDTKTTTGGGYGGGYGDSDTKTTTGGGYGGGYGDSDTKTTTGGGYGDD----RRE 718
Query: 151 EVDYKEEEKHHKKLEHLGELGTTAAGAYALHEKHKSEKDPENAHRHKLEEEVAAAAAVGA 210
+VDY++EEK H K EHLGELG AAG +AL+EKHKSEKDPENAHRHK+EEEVAA AAVG+
Sbjct: 719 DVDYEKEEKRHSKHEHLGELGAAAAGGFALYEKHKSEKDPENAHRHKIEEEVAAVAAVGS 898
Query: 211 GGFAFHEHHEKKEAKEEEEESHGKKHHHLFG 241
GGFAFHEHHEKKE KEEEEE+HGKK HH FG
Sbjct: 899 GGFAFHEHHEKKETKEEEEEAHGKKKHHFFG 991
>AW695835 similar to GP|14582465|gb| putative transcription factor {Vitis
vinifera}, partial (57%)
Length = 442
Score = 151 bits (381), Expect = 3e-37
Identities = 67/88 (76%), Positives = 79/88 (89%)
Frame = -2
Query: 155 KEEEKHHKKLEHLGELGTTAAGAYALHEKHKSEKDPENAHRHKLEEEVAAAAAVGAGGFA 214
K E HK LEHLGELGT AAG YA+HEKH+++KDPE+AH+HK+EEE+AAAAAVG+GGF
Sbjct: 405 KRGEHIHKHLEHLGELGTAAAGVYAMHEKHEAKKDPEHAHKHKIEEEIAAAAAVGSGGFV 226
Query: 215 FHEHHEKKEAKEEEEESHGKKHHHLFGR 242
FHEHHEKKEAK+E+EE+HGKKHHHLF R
Sbjct: 225 FHEHHEKKEAKKEDEEAHGKKHHHLF*R 142
>AL372330 similar to GP|21322752|dbj cold shock protein-1 {Triticum
aestivum}, partial (33%)
Length = 435
Score = 81.3 bits (199), Expect = 3e-16
Identities = 53/107 (49%), Positives = 58/107 (53%), Gaps = 6/107 (5%)
Frame = +2
Query: 46 YKKSSYETSGGGYETGYNKTSSYSTDEPNSGGG---GYDSGYNKTAYSTDEPSGGGYGTG 102
Y+ SS GG GY +SS S +SGGG G GY ++ S GGYG G
Sbjct: 5 YRSSSSGGYGGSSSGGYGGSSSSSYPASSSGGGYGGGSSGGYGGSSGGYGG-SSGGYGGG 181
Query: 103 GAGGGYSTG-GGYGTGGGGGGEYSTGG-GGGGY-STGGGGYGNDDSG 146
G GGG S G GG G GGG GG Y GG GGGGY GGGGYG D G
Sbjct: 182 GYGGGASGGYGGGGYGGGSGGGYGGGGYGGGGYGGGGGGGYGGDKMG 322
>TC76704 homologue to SP|Q09134|GRPA_MEDFA Abscisic acid and environmental
stress inducible protein. [Sickle medic] {Medicago
falcata}, partial (81%)
Length = 771
Score = 80.5 bits (197), Expect = 5e-16
Identities = 47/98 (47%), Positives = 54/98 (54%), Gaps = 6/98 (6%)
Frame = +2
Query: 55 GGGYETG-YNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGG 113
GGGY G YN Y+ N GGGGY++G GGY GG GGY+ GGG
Sbjct: 191 GGGYNGGGYNHGGGYNGGGYNHGGGGYNNG-------------GGYNHGG--GGYNNGGG 325
Query: 114 YGTGGGG--GGEYSTGGG---GGGYSTGGGGYGNDDSG 146
Y GGGG GG Y+ GGG GGGY+ GGGGY + G
Sbjct: 326 YNHGGGGYNGGGYNHGGGGYNGGGYNHGGGGYNHGGGG 439
Score = 62.8 bits (151), Expect = 1e-10
Identities = 43/108 (39%), Positives = 52/108 (47%), Gaps = 8/108 (7%)
Frame = +2
Query: 29 KTSYSTDEPSGGDYDSGYKKSSYETSGGGYETG---YNKTSSYSTDEPNSGGGGYDSG-- 83
KT+ D GG Y+ G +GGGY G YN Y N GGGGY++G
Sbjct: 161 KTNEVNDAKYGGGYNGGGYNHGGGYNGGGYNHGGGGYNNGGGY-----NHGGGGYNNGGG 325
Query: 84 YNKTAYSTDEPSGGGYGTGGAG---GGYSTGGGYGTGGGGGGEYSTGG 128
YN + +GGGY GG G GGY+ GGG GGGG +Y G
Sbjct: 326 YN---HGGGGYNGGGYNHGGGGYNGGGYNHGGGGYNHGGGGCQYHCHG 460
Score = 62.4 bits (150), Expect = 2e-10
Identities = 36/75 (48%), Positives = 39/75 (52%), Gaps = 20/75 (26%)
Frame = +2
Query: 86 KTAYSTDEPSGGGYGTGG--------------AGGGYSTGGGYGTGGGG---GGEYSTGG 128
KT D GGGY GG GGGY+ GGGY GGGG GG Y+ GG
Sbjct: 161 KTNEVNDAKYGGGYNGGGYNHGGGYNGGGYNHGGGGYNNGGGYNHGGGGYNNGGGYNHGG 340
Query: 129 G---GGGYSTGGGGY 140
G GGGY+ GGGGY
Sbjct: 341 GGYNGGGYNHGGGGY 385
Score = 61.6 bits (148), Expect = 3e-10
Identities = 41/87 (47%), Positives = 47/87 (53%), Gaps = 10/87 (11%)
Frame = +2
Query: 70 TDEPNSG--GGGYDSG-YNKTAYSTDEPSGGGYGTGG---AGGGYSTGGGYGTGGGGGGE 123
T+E N GGGY+ G YN GGGY GG GGGY+ GGGY GGGG
Sbjct: 164 TNEVNDAKYGGGYNGGGYNH---------GGGYNGGGYNHGGGGYNNGGGYNHGGGG--- 307
Query: 124 YSTGGG----GGGYSTGGGGYGNDDSG 146
Y+ GGG GGGY+ GGGY + G
Sbjct: 308 YNNGGGYNHGGGGYN--GGGYNHGGGG 382
Score = 56.6 bits (135), Expect = 8e-09
Identities = 30/59 (50%), Positives = 35/59 (58%), Gaps = 2/59 (3%)
Frame = +2
Query: 91 TDEPSGGGYGTGGAGGGYSTGGGYGTGG--GGGGEYSTGGGGGGYSTGGGGYGNDDSGN 147
T+E + YG G GGGY+ GGGY GG GGG Y+ GGG Y+ GGGGY N N
Sbjct: 164 TNEVNDAKYGGGYNGGGYNHGGGYNGGGYNHGGGGYNNGGG---YNHGGGGYNNGGGYN 331
Score = 36.6 bits (83), Expect = 0.009
Identities = 28/88 (31%), Positives = 32/88 (35%)
Frame = +2
Query: 26 GYNKTSYSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYN 85
GYN G + GY +GGGY N GGGGY
Sbjct: 266 GYNNGGGYNHGGGGYNNGGGYNHGGGGYNGGGY---------------NHGGGGY----- 385
Query: 86 KTAYSTDEPSGGGYGTGGAGGGYSTGGG 113
+GGGY G GGGY+ GGG
Sbjct: 386 ---------NGGGYNHG--GGGYNHGGG 436
>TC76707 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulated protein
CORA. [Alfalfa] {Medicago sativa}, partial (87%)
Length = 780
Score = 77.0 bits (188), Expect = 6e-15
Identities = 54/147 (36%), Positives = 70/147 (46%), Gaps = 8/147 (5%)
Frame = +1
Query: 21 TDTETGYNKTSYSTDEPSGGDYDSGYKKSSYETSGGGYETG---YNKTSSYSTDEPNSGG 77
TDT+ + + ++ G Y GY +GGGY G YN N GG
Sbjct: 100 TDTKKEVVEKTNEVNDAKYGGYGGGYNHGGGGYNGGGYNHGGGGYNNGGGGY----NHGG 267
Query: 78 GGYDSGYNKTAYSTDEPSGGGYGTGGAG-----GGYSTGGGYGTGGGGGGEYSTGGGGGG 132
GGY++G + +GGGY GG G GGY+ GGG GGGG G G GGGG
Sbjct: 268 GGYNNGGGGYNHGGGGYNGGGYNHGGGGYNHGGGGYNHGGGGYNGGGGHG----GHGGGG 435
Query: 133 YSTGGGGYGNDDSGNRRDEVDYKEEEK 159
Y+ GGGG+G + + V + EEK
Sbjct: 436 YN-GGGGHGGHGAA---ESVAVQTEEK 504
Score = 76.6 bits (187), Expect = 8e-15
Identities = 58/169 (34%), Positives = 72/169 (42%), Gaps = 35/169 (20%)
Frame = +1
Query: 26 GYNKTSYSTDEPSGGDYDSGYKKSSYETSGGGYETG---YNKTSS-YSTDEPNSGGGGYD 81
GYN Y+ GG Y++G Y GGGY G YN Y+ N GGGGY+
Sbjct: 193 GYNGGGYNH---GGGGYNNG--GGGYNHGGGGYNNGGGGYNHGGGGYNGGGYNHGGGGYN 357
Query: 82 SGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGT-----------------------GG 118
G + +GGG G GGGY+ GGG+G GG
Sbjct: 358 HGGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGG 537
Query: 119 G---GGGEYSTGGG-----GGGYSTGGGGYGNDDSGNRRDEVDYKEEEK 159
G GGG Y+ GGG GGGY+ GGGG+G G + E+K
Sbjct: 538 GYNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGGGGHGGHGAEQTEDK 684
Score = 42.0 bits (97), Expect = 2e-04
Identities = 35/100 (35%), Positives = 43/100 (43%), Gaps = 11/100 (11%)
Frame = +1
Query: 38 SGGDYDSGYKKSSYETSGGGYETGYNKTSSYS------TDEPNSG--GGGYDSG---YNK 86
+GG G+ Y +GGG G+ S + T+E N GGGY+ G YN
Sbjct: 397 NGGGGHGGHGGGGY--NGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGGSYNH 570
Query: 87 TAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYST 126
S GGGY GG G G GGG+G G E T
Sbjct: 571 GGGSYHH-GGGGYNHGGGGHGGHGGGGHGGHGAEQTEDKT 687
>TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 912
Score = 72.8 bits (177), Expect = 1e-13
Identities = 52/155 (33%), Positives = 64/155 (40%), Gaps = 15/155 (9%)
Frame = +1
Query: 7 DRGLFHHHK--NEDRPTDTETGYNKTSYSTDEPSGGDYDSGYKKSSYETSGGGYETGYNK 64
D + H H+ DR T G+ +++ ++ + E G G N
Sbjct: 322 DSFITHSHQ*IINDRETGRSRGFGFVTFANEKSMN---------DAIEAMNGQDLDGRNI 474
Query: 65 TSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAG-----GGYSTGGGYGTGGG 119
T + + + GGGG G GGGYG GG G GY GGGYG GGG
Sbjct: 475 TVNQAQSRGSGGGGGGGRG------------GGGYGGGGGGYGGERRGYGGGGGYGGGGG 618
Query: 120 GG--------GEYSTGGGGGGYSTGGGGYGNDDSG 146
GG G YS GGGGGGY GG G D G
Sbjct: 619 GGYGERRGGGGGYSRGGGGGGYGGGGYSRGGGDGG 723
Score = 63.2 bits (152), Expect = 9e-11
Identities = 44/100 (44%), Positives = 44/100 (44%), Gaps = 4/100 (4%)
Frame = +1
Query: 38 SGGDYDSGYKKSSYETSGGGY---ETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEP 94
SGG G Y GGGY GY GGGGY G
Sbjct: 502 SGGGGGGGRGGGGYGGGGGGYGGERRGYG------------GGGGYGGG----------- 612
Query: 95 SGGGYGTG-GAGGGYSTGGGYGTGGGGGGEYSTGGGGGGY 133
GGGYG G GGGYS GGG GG GGG YS GGG GGY
Sbjct: 613 GGGGYGERRGGGGGYSRGGG--GGGYGGGGYSRGGGDGGY 726
>TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 1054
Score = 72.0 bits (175), Expect = 2e-13
Identities = 49/142 (34%), Positives = 59/142 (41%), Gaps = 13/142 (9%)
Frame = +1
Query: 18 DRPTDTETGYNKTSYSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGG 77
DR T G+ +++ ++ + E G G N T + + + GG
Sbjct: 457 DRETGRSRGFGFVTFANEKSMN---------DAIEAMNGQDLDGRNITVNQAQSRGSGGG 609
Query: 78 GGYDSGYNKTAYSTDEPSGGGYGTGGAG-----GGYSTGGGYGTGGGGG--------GEY 124
GG G GGGYG GG G GY GGGYG GGGGG G Y
Sbjct: 610 GGGGRG------------GGGYGGGGGGYGGERRGYGGGGGYGGGGGGGYGERRGGGGGY 753
Query: 125 STGGGGGGYSTGGGGYGNDDSG 146
S GGGGGGY GG G D G
Sbjct: 754 SRGGGGGGYGGGGYSRGGGDGG 819
Score = 63.2 bits (152), Expect = 9e-11
Identities = 44/100 (44%), Positives = 44/100 (44%), Gaps = 4/100 (4%)
Frame = +1
Query: 38 SGGDYDSGYKKSSYETSGGGY---ETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEP 94
SGG G Y GGGY GY GGGGY G
Sbjct: 598 SGGGGGGGRGGGGYGGGGGGYGGERRGYG------------GGGGYGGG----------- 708
Query: 95 SGGGYGTG-GAGGGYSTGGGYGTGGGGGGEYSTGGGGGGY 133
GGGYG G GGGYS GGG GG GGG YS GGG GGY
Sbjct: 709 GGGGYGERRGGGGGYSRGGG--GGGYGGGGYSRGGGDGGY 822
>TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 754
Score = 72.0 bits (175), Expect = 2e-13
Identities = 49/142 (34%), Positives = 59/142 (41%), Gaps = 13/142 (9%)
Frame = +2
Query: 18 DRPTDTETGYNKTSYSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGG 77
DR T G+ +++ ++ + E G G N T + + + GG
Sbjct: 203 DRETGRSRGFGFVTFANEKSMN---------DAIEAMNGQDLDGRNITVNQAQSRGSGGG 355
Query: 78 GGYDSGYNKTAYSTDEPSGGGYGTGGAG-----GGYSTGGGYGTGGGGG--------GEY 124
GG G GGGYG GG G GY GGGYG GGGGG G Y
Sbjct: 356 GGGGRG------------GGGYGGGGGGYGGERRGYGGGGGYGGGGGGGYGERRGGGGGY 499
Query: 125 STGGGGGGYSTGGGGYGNDDSG 146
S GGGGGGY GG G D G
Sbjct: 500 SRGGGGGGYGGGGYSRGGGDGG 565
Score = 63.2 bits (152), Expect = 9e-11
Identities = 44/100 (44%), Positives = 44/100 (44%), Gaps = 4/100 (4%)
Frame = +2
Query: 38 SGGDYDSGYKKSSYETSGGGY---ETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEP 94
SGG G Y GGGY GY GGGGY G
Sbjct: 344 SGGGGGGGRGGGGYGGGGGGYGGERRGYG------------GGGGYGGG----------- 454
Query: 95 SGGGYGTG-GAGGGYSTGGGYGTGGGGGGEYSTGGGGGGY 133
GGGYG G GGGYS GGG GG GGG YS GGG GGY
Sbjct: 455 GGGGYGERRGGGGGYSRGGG--GGGYGGGGYSRGGGDGGY 568
>TC76706 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulated protein
CORA. [Alfalfa] {Medicago sativa}, partial (94%)
Length = 1034
Score = 71.2 bits (173), Expect = 3e-13
Identities = 53/140 (37%), Positives = 67/140 (47%), Gaps = 1/140 (0%)
Frame = +1
Query: 21 TDTETGYNKTSYS-TDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGG 79
T+T T K T+E + Y GY Y GG GYN Y ++GGGG
Sbjct: 118 TETSTDAKKEVVEKTNEVNDAKY-GGYNHGGYNHGGGYNGGGYNHGGGY-----HNGGGG 279
Query: 80 YDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGG 139
Y +G GGGY GG GGY+ GGG+G GG Y+ GGG GGY+ GGGG
Sbjct: 280 YHNG------------GGGYNHGG--GGYNGGGGHGGHGG----YNGGGGHGGYN-GGGG 402
Query: 140 YGNDDSGNRRDEVDYKEEEK 159
+G + + V + EEK
Sbjct: 403 HGGHGAA---ESVAVQTEEK 453
Score = 70.9 bits (172), Expect = 4e-13
Identities = 55/164 (33%), Positives = 68/164 (40%), Gaps = 30/164 (18%)
Frame = +1
Query: 26 GYNKTSYSTDEP-SGGDYDSGYKKSSYETSGGGYETG---YNKTSS-YSTDEPNSGGGGY 80
GYN Y+ +GG Y+ G Y GGGY G YN Y+ + G GGY
Sbjct: 190 GYNHGGYNHGGGYNGGGYNHG---GGYHNGGGGYHNGGGGYNHGGGGYNGGGGHGGHGGY 360
Query: 81 DSGYNKTAYSTDEPSGGGYGTGGA---------------------GGGYSTGGGYGTGGG 119
+ G Y+ GGG+G GA GG Y+ GG Y GGG
Sbjct: 361 NGGGGHGGYN----GGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGGSYNHGGSYNHGGG 528
Query: 120 ----GGGEYSTGGGGGGYSTGGGGYGNDDSGNRRDEVDYKEEEK 159
GGG Y GGGGY+ GGGG+G G + E+K
Sbjct: 529 SYNHGGGSYH--HGGGGYNHGGGGHGGHGGGGHGGHGAEQTEDK 654
Score = 40.8 bits (94), Expect = 5e-04
Identities = 32/102 (31%), Positives = 41/102 (39%), Gaps = 15/102 (14%)
Frame = +1
Query: 40 GDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNS-------GGGGYDSG--YNKTAYS 90
G Y+ G Y GG G ++ + T+E + GGG Y+ G YN S
Sbjct: 352 GGYNGGGGHGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGGSYNHGGSYNHGGGS 531
Query: 91 TDEPSG------GGYGTGGAGGGYSTGGGYGTGGGGGGEYST 126
+ G GGY GG G G GGG+G G E T
Sbjct: 532 YNHGGGSYHHGGGGYNHGGGGHGGHGGGGHGGHGAEQTEDKT 657
>BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melanogaster},
partial (26%)
Length = 308
Score = 69.3 bits (168), Expect = 1e-12
Identities = 39/82 (47%), Positives = 40/82 (48%)
Frame = +2
Query: 65 TSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEY 124
T S + P GGGG G GGG G GG GGG GGG G GGGGGG
Sbjct: 8 TIPLSGENPGGGGGGGGGGGGGGG------GGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 169
Query: 125 STGGGGGGYSTGGGGYGNDDSG 146
GGGGGG GGGG G G
Sbjct: 170 GGGGGGGGGGGGGGGGGGGGGG 235
Score = 67.4 bits (163), Expect = 5e-12
Identities = 36/71 (50%), Positives = 36/71 (50%)
Frame = +3
Query: 76 GGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYST 135
GGGG G GGG G GG GGG GGG G GGGGGG GGGGGG
Sbjct: 51 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 230
Query: 136 GGGGYGNDDSG 146
GGGG G G
Sbjct: 231 GGGGGGGGGGG 263
Score = 67.4 bits (163), Expect = 5e-12
Identities = 36/71 (50%), Positives = 36/71 (50%)
Frame = +3
Query: 76 GGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYST 135
GGGG G GGG G GG GGG GGG G GGGGGG GGGGGG
Sbjct: 93 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 272
Query: 136 GGGGYGNDDSG 146
GGGG G G
Sbjct: 273 GGGGGGGGGGG 305
Score = 67.4 bits (163), Expect = 5e-12
Identities = 36/71 (50%), Positives = 36/71 (50%)
Frame = +3
Query: 76 GGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYST 135
GGGG G GGG G GG GGG GGG G GGGGGG GGGGGG
Sbjct: 66 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 245
Query: 136 GGGGYGNDDSG 146
GGGG G G
Sbjct: 246 GGGGGGGGGGG 278
Score = 67.4 bits (163), Expect = 5e-12
Identities = 36/71 (50%), Positives = 36/71 (50%)
Frame = +3
Query: 76 GGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYST 135
GGGG G GGG G GG GGG GGG G GGGGGG GGGGGG
Sbjct: 72 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 251
Query: 136 GGGGYGNDDSG 146
GGGG G G
Sbjct: 252 GGGGGGGGGGG 284
Score = 67.4 bits (163), Expect = 5e-12
Identities = 36/71 (50%), Positives = 36/71 (50%)
Frame = +1
Query: 76 GGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYST 135
GGGG G GGG G GG GGG GGG G GGGGGG GGGGGG
Sbjct: 73 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 252
Query: 136 GGGGYGNDDSG 146
GGGG G G
Sbjct: 253 GGGGGGGGGGG 285
Score = 67.4 bits (163), Expect = 5e-12
Identities = 36/71 (50%), Positives = 36/71 (50%)
Frame = +1
Query: 76 GGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYST 135
GGGG G GGG G GG GGG GGG G GGGGGG GGGGGG
Sbjct: 55 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 234
Query: 136 GGGGYGNDDSG 146
GGGG G G
Sbjct: 235 GGGGGGGGGGG 267
Score = 67.4 bits (163), Expect = 5e-12
Identities = 36/71 (50%), Positives = 36/71 (50%)
Frame = +1
Query: 76 GGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYST 135
GGGG G GGG G GG GGG GGG G GGGGGG GGGGGG
Sbjct: 94 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 273
Query: 136 GGGGYGNDDSG 146
GGGG G G
Sbjct: 274 GGGGGGGGGGG 306
Score = 67.0 bits (162), Expect = 6e-12
Identities = 38/81 (46%), Positives = 39/81 (47%)
Frame = +3
Query: 66 SSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYS 125
S + P GGGG G GGG G GG GGG GGG G GGGGGG
Sbjct: 12 SPFPGKTPGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG---GGGGGGGGGGGGGGG 182
Query: 126 TGGGGGGYSTGGGGYGNDDSG 146
GGGGGG GGGG G G
Sbjct: 183 GGGGGGGGGGGGGGGGGGGGG 245
>TC89001 similar to GP|5726567|gb|AAD48471.1| glycine-rich RNA-binding
protein {Glycine max}, partial (90%)
Length = 488
Score = 68.6 bits (166), Expect = 2e-12
Identities = 48/132 (36%), Positives = 56/132 (42%), Gaps = 9/132 (6%)
Frame = +3
Query: 24 ETGYNKTSYSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSG 83
E ++K TD D ++G + G G+ T + S E +G G
Sbjct: 102 EQAFSKFGEITDSKVINDRETGRSR------GFGFVTFAEEKSMRDAIEEMNGQD--IDG 257
Query: 84 YNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGG--------GGEYSTGGGGGGYST 135
N T GG G GG GGGY GGGYG GGGG G YS GGGGGY
Sbjct: 258 RNITVNEAQSRGSGGGGRGGGGGGYGGGGGYGGGGGGYGGGGGRRDGGYSRSGGGGGYGG 437
Query: 136 GGG-GYGNDDSG 146
GG GYG G
Sbjct: 438 GGDRGYGGGGGG 473
Score = 60.8 bits (146), Expect = 4e-10
Identities = 44/130 (33%), Positives = 55/130 (41%), Gaps = 6/130 (4%)
Frame = +3
Query: 18 DRPTDTETGYNKTSYSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGG 77
DR T G+ +++ ++ + + E G G N T + + + GG
Sbjct: 153 DRETGRSRGFGFVTFAEEKSM---------RDAIEEMNGQDIDGRNITVNEAQSRGSGGG 305
Query: 78 G--GYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGG----GGGGEYSTGGGGG 131
G G GY GGG G GG GGGY GGG GG GGGG Y GG G
Sbjct: 306 GRGGGGGGY-----------GGGGGYGGGGGGYGGGGGRRDGGYSRSGGGGGYGGGGDRG 452
Query: 132 GYSTGGGGYG 141
GGGGYG
Sbjct: 453 YGGGGGGGYG 482
Score = 48.5 bits (114), Expect = 2e-06
Identities = 25/39 (64%), Positives = 25/39 (64%)
Frame = +3
Query: 112 GGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGNRRD 150
GG G GGGGGG GGGGGY GGGGYG G RRD
Sbjct: 297 GGGGRGGGGGGY----GGGGGYGGGGGGYGG--GGGRRD 395
>BQ155184 homologue to GP|11994861|dbj putative ripening protein {Calystegia
soldanella}, partial (12%)
Length = 589
Score = 67.4 bits (163), Expect = 5e-12
Identities = 28/33 (84%), Positives = 29/33 (87%)
Frame = +2
Query: 209 GAGGFAFHEHHEKKEAKEEEEESHGKKHHHLFG 241
G GGFAFHEHHEKKE KEEEEE+HGKK HH FG
Sbjct: 437 GFGGFAFHEHHEKKETKEEEEEAHGKKKHHFFG 535
>TC85301 similar to GP|8096255|dbj|BAA96106.1 TrPRP2 {Trifolium repens},
partial (92%)
Length = 1318
Score = 65.5 bits (158), Expect = 2e-11
Identities = 46/113 (40%), Positives = 59/113 (51%), Gaps = 13/113 (11%)
Frame = -2
Query: 38 SGGDYDSGYKKSSYETSG---GGYETGYNKTSSYSTDEPNSGG----GGYDSGYNKTAYS 90
+GG Y G + T G GG+ TG T +ST +GG G Y G++ +S
Sbjct: 576 TGGLYTGGLYTGGFSTGGL*TGGFSTGGLYTGGFSTGGL*TGGFSTGGLYTGGFSTGGFS 397
Query: 91 TDEPSGGGYGTGGA-GGGYSTG----GGYGTGGGGGGEYSTGG-GGGGYSTGG 137
T GG+ TGG GG+STG GG+ TGG G +STGG GG+STGG
Sbjct: 396 TGGL*AGGFSTGGLYTGGFSTGGL*IGGFSTGGLYTGGFSTGGL*TGGFSTGG 238
Score = 59.7 bits (143), Expect = 1e-09
Identities = 49/120 (40%), Positives = 63/120 (51%), Gaps = 17/120 (14%)
Frame = -2
Query: 38 SGGDYDSGYKKSS-----YETSG---GGYETGYNKTSSYSTDEPNSGG---GGYDSGYNK 86
+GG Y G+ + T G GG+ TG T +ST +GG GG+ +G
Sbjct: 696 TGGLYTGGFSTGGL*IGGFSTGGLYTGGFSTGGL*TGGFSTGGLYTGGLYTGGFSTGGL* 517
Query: 87 TA-YSTDEPSGGGYGTGGA-GGGYSTGGGYGTGGGGGGEYSTGG-GGGGYSTGG---GGY 140
T +ST GG+ TGG GG+STGG Y TGG G +STGG GG+STGG GG+
Sbjct: 516 TGGFSTGGLYTGGFSTGGL*TGGFSTGGLY-TGGFSTGGFSTGGL*AGGFSTGGLYTGGF 340
Score = 57.8 bits (138), Expect = 4e-09
Identities = 45/113 (39%), Positives = 56/113 (48%), Gaps = 13/113 (11%)
Frame = -2
Query: 38 SGGDYDSGYKKSSYETSG---GGYETGYNKTSSYSTDEPNSGG----GGYDSGYNKTAYS 90
+GG Y G+ + T G GG+ TG T +ST GG G Y G++
Sbjct: 441 TGGLYTGGFSTGGFSTGGL*AGGFSTGGLYTGGFSTGGL*IGGFSTGGLYTGGFSTGGL* 262
Query: 91 TDEPSGGGYGTGGA-GGGYSTG----GGYGTGGGGGGEYSTGG-GGGGYSTGG 137
T S GG TGG GG+STG GG+ TGG G +STGG GG+ TGG
Sbjct: 261 TGGFSTGGLYTGGL*TGGFSTGGLYTGGFSTGGL*TGGFSTGGLYTGGW*TGG 103
Score = 57.8 bits (138), Expect = 4e-09
Identities = 50/121 (41%), Positives = 65/121 (53%), Gaps = 18/121 (14%)
Frame = -2
Query: 38 SGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGG---GGYDSGYNKTA-YSTDE 93
+GG Y G+ T GG+ TG T +ST ++GG GG+ +G T +ST
Sbjct: 501 TGGLYTGGFSTGGL*T--GGFSTGGLYTGGFSTGGFSTGGL*AGGFSTGGLYTGGFSTGG 328
Query: 94 PSGGGYGTGGA-GGGYSTG----GGYGTGG---GG--GGEYSTGG-GGGGYSTGG---GG 139
GG+ TGG GG+STG GG+ TGG GG G +STGG GG+STGG GG
Sbjct: 327 L*IGGFSTGGLYTGGFSTGGL*TGGFSTGGLYTGGL*TGGFSTGGLYTGGFSTGGL*TGG 148
Query: 140 Y 140
+
Sbjct: 147 F 145
Score = 57.4 bits (137), Expect = 5e-09
Identities = 48/119 (40%), Positives = 60/119 (50%), Gaps = 16/119 (13%)
Frame = -2
Query: 38 SGGDYDSGYKKSSYETSG---GGYETGYNKTSSYSTDEPNSGG----GGYDSGYNKTAYS 90
+GG Y G+ T G GG TG T +ST +GG G Y G++
Sbjct: 636 TGGLYTGGFSTGGL*TGGFSTGGLYTGGLYTGGFSTGGL*TGGFSTGGLYTGGFSTGGL* 457
Query: 91 TDEPSGGGYGTGG-AGGGYSTG----GGYGTGGGGGGEYSTGG-GGGGYSTGG---GGY 140
T S GG TGG + GG+STG GG+ TGG G +STGG GG+STGG GG+
Sbjct: 456 TGGFSTGGLYTGGFSTGGFSTGGL*AGGFSTGGLYTGGFSTGGL*IGGFSTGGLYTGGF 280
Score = 55.8 bits (133), Expect = 1e-08
Identities = 47/118 (39%), Positives = 58/118 (48%), Gaps = 16/118 (13%)
Frame = -2
Query: 39 GGDYDSGYKKSSYETSG---GGYETGYNKTSSYSTDEPNSGG----GGYDSGYNKTAYST 91
GG + G + T G GG+ TG T +ST GG G Y G++ T
Sbjct: 768 GGFSNGGL*TGGFSTGGL*TGGFSTGGLYTGGFSTGGL*IGGFSTGGLYTGGFSTGGL*T 589
Query: 92 DEPSGGGYGTGGA-GGGYSTGG----GYGTGGGGGGEYSTGGG-GGGYSTGG---GGY 140
S GG TGG GG+STGG G+ TGG G +STGG GG+STGG GG+
Sbjct: 588 GGFSTGGLYTGGLYTGGFSTGGL*TGGFSTGGLYTGGFSTGGL*TGGFSTGGLYTGGF 415
Score = 55.8 bits (133), Expect = 1e-08
Identities = 46/120 (38%), Positives = 58/120 (48%), Gaps = 17/120 (14%)
Frame = -2
Query: 38 SGGDYDSGYKKSSYETSG---GGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEP 94
+GG G + T G GG+ TG T +ST +GG Y Y+
Sbjct: 711 TGGFSTGGLYTGGFSTGGL*IGGFSTGGLYTGGFSTGGL*TGGFSTGGLYTGGLYT---- 544
Query: 95 SGGGYGTGGA-GGGYSTG----GGYGTGGGGGGEYSTGG---GG---GGYSTGG---GGY 140
GG+ TGG GG+STG GG+ TGG G +STGG GG GG+STGG GG+
Sbjct: 543 --GGFSTGGL*TGGFSTGGLYTGGFSTGGL*TGGFSTGGLYTGGFSTGGFSTGGL*AGGF 370
Score = 55.1 bits (131), Expect = 2e-08
Identities = 50/129 (38%), Positives = 63/129 (48%), Gaps = 26/129 (20%)
Frame = -2
Query: 38 SGGDYDSGYKKSSYETSG---GGYETGYNKTSSYSTDEPNSGG--------GGYDSGYNK 86
+GG + G K + T G GG+ G T YS GG GG+ +G
Sbjct: 891 NGGFSNGGL*KGGFSTGGL*MGGFSKGGLYTGGYSIGGL*IGGFSNGGL*TGGFSTGGL* 712
Query: 87 TA-YSTDEPSGGGYGTGGAG-GGYSTG----GGYGTGGGGGGEYSTGG---GG---GGYS 134
T +ST GG+ TGG GG+STG GG+ TGG G +STGG GG GG+S
Sbjct: 711 TGGFSTGGLYTGGFSTGGL*IGGFSTGGLYTGGFSTGGL*TGGFSTGGLYTGGLYTGGFS 532
Query: 135 TGG---GGY 140
TGG GG+
Sbjct: 531 TGGL*TGGF 505
Score = 53.5 bits (127), Expect = 7e-08
Identities = 47/120 (39%), Positives = 57/120 (47%), Gaps = 18/120 (15%)
Frame = -2
Query: 39 GGDYDSGYKKSSYETSG--------GGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTA-- 88
GG Y GY G GG+ TG T +ST GG Y G++
Sbjct: 813 GGLYTGGYSIGGL*IGGFSNGGL*TGGFSTGGL*TGGFST------GGLYTGGFSTGGL* 652
Query: 89 ---YSTDEPSGGGYGTGGA-GGGYSTGGGYGTGGGGGGEYSTGG-GGGGYSTGG---GGY 140
+ST GG+ TGG GG+STGG Y TGG G +STGG GG+STGG GG+
Sbjct: 651 IGGFSTGGLYTGGFSTGGL*TGGFSTGGLY-TGGLYTGGFSTGGL*TGGFSTGGLYTGGF 475
Score = 45.8 bits (107), Expect = 1e-05
Identities = 39/110 (35%), Positives = 48/110 (43%), Gaps = 7/110 (6%)
Frame = -2
Query: 38 SGGDYDSGYKKSSYETSG---GGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEP 94
+GG G + G GG+ TG +S GG Y GY+
Sbjct: 921 TGGSSTGGI*NGGFSNGGL*KGGFSTGGL*MGGFSK------GGLYTGGYSIGGL*IGGF 760
Query: 95 SGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGG-GGGGYSTGG---GGY 140
S GG TGG G GG+ TGG G +STGG GG+STGG GG+
Sbjct: 759 SNGGL*TGGFSTGGL*TGGFSTGGLYTGGFSTGGL*IGGFSTGGLYTGGF 610
Score = 45.1 bits (105), Expect = 3e-05
Identities = 39/109 (35%), Positives = 51/109 (46%), Gaps = 9/109 (8%)
Frame = -2
Query: 26 GYNKTSYSTDEP-SGGDYDSGYKKSSYETSG---GGYETGYNKTSSYSTDEPNSGG---- 77
G++ +ST +GG G + T G GG+ TG T +ST +GG
Sbjct: 420 GFSTGGFSTGGL*AGGFSTGGLYTGGFSTGGL*IGGFSTGGLYTGGFSTGGL*TGGFSTG 241
Query: 78 GGYDSGYNKTAYSTDEPSGGGYGTGGA-GGGYSTGGGYGTGGGGGGEYS 125
G Y G +ST GG+ TGG GG+STGG Y TGG G +S
Sbjct: 240 GLYTGGL*TGGFSTGGLYTGGFSTGGL*TGGFSTGGLY-TGGW*TGGFS 97
Score = 44.7 bits (104), Expect = 3e-05
Identities = 40/105 (38%), Positives = 52/105 (49%), Gaps = 18/105 (17%)
Frame = -2
Query: 54 SGGGYETGYNKTSSYSTDEPNSGG---GGYDSG------YNKTAYSTDEPSGGGYGTGGA 104
S GG TG + T ++GG GG+ +G ++K T GGY GG
Sbjct: 939 SNGGL*TGGSSTGGI*NGGFSNGGL*KGGFSTGGL*MGGFSKGGLYT-----GGYSIGGL 775
Query: 105 G-GGYSTGG----GYGTGGGGGGEYSTGG-GGGGYSTGG---GGY 140
GG+S GG G+ TGG G +STGG GG+STGG GG+
Sbjct: 774 *IGGFSNGGL*TGGFSTGGL*TGGFSTGGLYTGGFSTGGL*IGGF 640
Score = 44.3 bits (103), Expect = 4e-05
Identities = 38/94 (40%), Positives = 47/94 (49%), Gaps = 9/94 (9%)
Frame = -2
Query: 56 GGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGA-GGGYSTGG-- 112
GG+ G T ST +GG + G K +ST GG+ GG GGYS GG
Sbjct: 948 GGFSNGGL*TGGSSTGGI*NGGFS-NGGL*KGGFSTGGL*MGGFSKGGLYTGGYSIGGL* 772
Query: 113 --GYGTGGGGGGEYSTGGG-GGGYSTGG---GGY 140
G+ GG G +STGG GG+STGG GG+
Sbjct: 771 IGGFSNGGL*TGGFSTGGL*TGGFSTGGLYTGGF 670
Score = 39.7 bits (91), Expect = 0.001
Identities = 27/55 (49%), Positives = 31/55 (56%), Gaps = 9/55 (16%)
Frame = -2
Query: 97 GGYGTGGA-GGGYSTGG----GYGTGGGGGGEYSTGG-GGGGYSTGG---GGYGN 142
GG TGG GG+S GG G+ TGG G +S GG GGYS GG GG+ N
Sbjct: 918 GGSSTGGI*NGGFSNGGL*KGGFSTGGL*MGGFSKGGLYTGGYSIGGL*IGGFSN 754
>BM814132 weakly similar to OMNI|MT3615.1 PE_PGRS family protein
{Mycobacterium tuberculosis CDC1551}, partial (7%)
Length = 164
Score = 65.1 bits (157), Expect = 2e-11
Identities = 31/51 (60%), Positives = 31/51 (60%)
Frame = +3
Query: 96 GGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSG 146
GGG G GG GGG GGG G GGGGGG GGGGGG GGGG G G
Sbjct: 3 GGGRGGGGGGGGGGGGGGGGAGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 155
Score = 64.7 bits (156), Expect = 3e-11
Identities = 31/51 (60%), Positives = 32/51 (61%)
Frame = +1
Query: 96 GGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSG 146
GGG G GG GGG GGG G GGGGGG + GGGGGG GGGG G G
Sbjct: 10 GGGGGGGGGGGGGGGGGGRGGGGGGGGGGAGGGGGGGGGGGGGGGGGGGGG 162
Score = 63.2 bits (152), Expect = 9e-11
Identities = 30/47 (63%), Positives = 31/47 (65%)
Frame = +2
Query: 95 SGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYG 141
+GGG G GG GGG GGG G GGGGGG GGGGGG GGGG G
Sbjct: 11 AGGGGGGGGGGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGGGGGGGG 151
Score = 62.0 bits (149), Expect = 2e-10
Identities = 30/51 (58%), Positives = 30/51 (58%)
Frame = +1
Query: 96 GGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSG 146
GGG G GG GGG GGG G GGGGGG GGGGG GGGG G G
Sbjct: 4 GGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGAGGGGGGGGGGGGGGGGGGG 156
Score = 62.0 bits (149), Expect = 2e-10
Identities = 30/51 (58%), Positives = 30/51 (58%)
Frame = +2
Query: 96 GGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSG 146
G G G GG GGG GGG G GGGGGG GGGGGG GGGG G G
Sbjct: 8 GAGGGGGGGGGGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGGGGGGGGGGG 160
Score = 59.7 bits (143), Expect = 1e-09
Identities = 31/53 (58%), Positives = 31/53 (58%), Gaps = 2/53 (3%)
Frame = +1
Query: 96 GGGYGTGGAGGGYSTGGGYG--TGGGGGGEYSTGGGGGGYSTGGGGYGNDDSG 146
GGG G GG GGG GGG G GGGGGG GGGGGG GGGG G G
Sbjct: 1 GGGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGAGGGGGGGGGGGGGGGGGGGG 159
Score = 52.8 bits (125), Expect = 1e-07
Identities = 30/57 (52%), Positives = 31/57 (53%)
Frame = +3
Query: 76 GGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGG 132
GGGG G +GGG G GG GGG GGG G GGGGGG GGGGGG
Sbjct: 39 GGGGGGGG-----------AGGGGGGGGGGGGGGGGGGGGGGGGGGG----GGGGGG 164
Score = 50.8 bits (120), Expect = 5e-07
Identities = 30/56 (53%), Positives = 30/56 (53%)
Frame = +3
Query: 76 GGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGG 131
GGGG G A GGG G GG GGG GGG G GGGGGG GGGGG
Sbjct: 27 GGGGGGGGGGGGA------GGGGGGGGGGGGGGGGGGGGGGGGGGGG----GGGGG 164
Score = 49.7 bits (117), Expect = 1e-06
Identities = 28/54 (51%), Positives = 28/54 (51%)
Frame = +1
Query: 76 GGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGG 129
GGGG G GGG G GGAGGG GGG G GGGGGG GGG
Sbjct: 13 GGGGGGGGGGGGGGGGGRGGGGGGGGGGAGGGGGGGGGGGGGGGGGG----GGG 162
Score = 43.5 bits (101), Expect = 7e-05
Identities = 22/38 (57%), Positives = 22/38 (57%)
Frame = +2
Query: 111 GGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGNR 148
GGG G GGGGGG GGGGG GGGG G G R
Sbjct: 2 GGGAGGGGGGGG-----GGGGGGGGGGGGGGGGGGGGR 100
Score = 41.2 bits (95), Expect = 4e-04
Identities = 21/37 (56%), Positives = 21/37 (56%)
Frame = +2
Query: 113 GYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGNRR 149
G G GGGGGG GGGGGG GGGG G G R
Sbjct: 2 GGGAGGGGGG----GGGGGGGGGGGGGGGGGGGGGGR 100
>TC76705 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulated protein
CORA. [Alfalfa] {Medicago sativa}, partial (80%)
Length = 854
Score = 64.7 bits (156), Expect = 3e-11
Identities = 53/170 (31%), Positives = 66/170 (38%), Gaps = 39/170 (22%)
Frame = +2
Query: 21 TDTETGYNKTSYS-TDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEP-NSGGG 78
T+T T K T+E + Y GY Y GG GYN Y ++GGG
Sbjct: 128 TETSTDAKKEVVEKTNEVNDAKY-GGYNHGGYNHGGGYNGGGYNHGGGYHNGGGYHNGGG 304
Query: 79 GYDSGYNKTAYSTDEPSGGGYGTG---GAGGGYSTGGGYG-------------------- 115
GY +G GGGY G G GGY+ GGG+G
Sbjct: 305 GYHNG------------GGGYNGGGGHGGHGGYNRGGGHGGRGAAESVAVQTEEKTNEVN 448
Query: 116 --TGGGGGGEYSTGG------------GGGGYSTGGGGYGNDDSGNRRDE 151
GGGGG ++ GG G GGY+ GGGG+G G+ E
Sbjct: 449 DARNGGGGGSFNKGGASYNHGRGSYHHG*GGYNHGGGGHGGGGHGSHGAE 598
Score = 55.5 bits (132), Expect = 2e-08
Identities = 35/89 (39%), Positives = 42/89 (46%), Gaps = 4/89 (4%)
Frame = +2
Query: 74 NSGGGGYDSGYNKTAYSTDEPSGGGYGTGGA----GGGYSTGGGYGTGGGGGGEYSTGGG 129
N GG + GYN Y+ GGGY GG GGGY GGG GGGG GG
Sbjct: 206 NHGGYNHGGGYNGGGYN----HGGGYHNGGGYHNGGGGYHNGGGGYNGGGG------HGG 355
Query: 130 GGGYSTGGGGYGNDDSGNRRDEVDYKEEE 158
GGY+ GGG G + + + + K E
Sbjct: 356 HGGYNRGGGHGGRGAAESVAVQTEEKTNE 442
Score = 52.0 bits (123), Expect = 2e-07
Identities = 45/145 (31%), Positives = 59/145 (40%), Gaps = 23/145 (15%)
Frame = +2
Query: 26 GYNKTSYSTDEPSGGDYDSG-YKKSSYETSGGGYETG----YNKTSSYSTDEPNSGGGGY 80
GYN Y+ GG Y+ G Y +GGGY G +N Y+ + G GGY
Sbjct: 200 GYNHGGYN----HGGGYNGGGYNHGGGYHNGGGYHNGGGGYHNGGGGYNGGGGHGGHGGY 367
Query: 81 DSG----------------YNKTAYSTDEPSGGGYGTGGAGGG-YSTGGGYGTGGGGGGE 123
+ G KT D +GGG G+ GG Y+ G G G GG
Sbjct: 368 NRGGGHGGRGAAESVAVQTEEKTNEVNDARNGGGGGSFNKGGASYNHGRGSYHHG*GGYN 547
Query: 124 YSTGG-GGGGYSTGGGGYGNDDSGN 147
+ GG GGGG+ + G D + N
Sbjct: 548 HGGGGHGGGGHGSHGAEQTEDKTQN 622
>TC87796 weakly similar to SP|P03211|EBN1_EBV EBNA-1 nuclear protein.
[strain B95-8 Human herpesvirus 4] {Epstein-barr
virus}, partial (23%)
Length = 431
Score = 64.3 bits (155), Expect = 4e-11
Identities = 40/89 (44%), Positives = 46/89 (50%), Gaps = 2/89 (2%)
Frame = +1
Query: 55 GGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGY 114
GGGY G GGGG G + Y + +GGGYG G GGG GGG
Sbjct: 13 GGGYGGGGGS---------GGGGGGGAGGAHGVGYGSGGGTGGGYGGGSPGGG---GGGG 156
Query: 115 GTGGGGGGEYSTGGG-GGGYSTG-GGGYG 141
G+GGGGGG + GG GGG G GGG+G
Sbjct: 157 GSGGGGGGGGAHGGAYGGGIGGGEGGGHG 243
Score = 61.6 bits (148), Expect = 3e-10
Identities = 37/77 (48%), Positives = 38/77 (49%), Gaps = 7/77 (9%)
Frame = +1
Query: 77 GGGYDSGYNKTAYSTDEPSGGGYGTGGA-----GGGYSTGGGYGTG--GGGGGEYSTGGG 129
GGGY G GGG G GGA G G TGGGYG G GGGGG +GGG
Sbjct: 13 GGGYGGGGGSGG-------GGGGGAGGAHGVGYGSGGGTGGGYGGGSPGGGGGGGGSGGG 171
Query: 130 GGGYSTGGGGYGNDDSG 146
GGG GG YG G
Sbjct: 172 GGGGGAHGGAYGGGIGG 222
Score = 53.9 bits (128), Expect = 5e-08
Identities = 37/99 (37%), Positives = 41/99 (41%)
Frame = +1
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
G D+ GY GGG G Y + GGGY G P GGG
Sbjct: 1 GRDHGGGYGGGGGSGGGGGGGAGGAHGVGYGSG--GGTGGGYGGG---------SPGGGG 147
Query: 99 YGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGG 137
G GG+GGG GG +G GGG GGG GGY G
Sbjct: 148 -GGGGSGGGGGGGGAHGGAYGGGIGGGEGGGHGGYEP*G 261
Score = 40.8 bits (94), Expect = 5e-04
Identities = 24/46 (52%), Positives = 25/46 (54%), Gaps = 6/46 (13%)
Frame = +1
Query: 107 GYSTGGGYGTGG--GGGGEYSTGGGGG-GYSTG---GGGYGNDDSG 146
G GGGYG GG GGGG GG G GY +G GGGYG G
Sbjct: 1 GRDHGGGYGGGGGSGGGGGGGAGGAHGVGYGSGGGTGGGYGGGSPG 138
>TC85302 similar to GP|437310|gb|AAA62850.1|| nodulin {Medicago truncatula},
partial (72%)
Length = 907
Score = 62.0 bits (149), Expect = 2e-10
Identities = 47/113 (41%), Positives = 60/113 (52%), Gaps = 13/113 (11%)
Frame = -1
Query: 38 SGGDYDSGYKKSSYETSG---GGYETGYNKTSSYSTDEPNSGG---GGYDSGYNKTA-YS 90
+GG Y G+ + T G GG+ TG T +ST GG GG+ +G T +S
Sbjct: 787 TGGLYTGGFSTGGFSTGGL*AGGFSTGGLYTGGFST-----GGL*IGGFSTGGLYTGGFS 623
Query: 91 TDEPSGGGYGTGGA-GGGYSTGG----GYGTGGGGGGEYSTGG-GGGGYSTGG 137
T GG+ TGG GG+STGG G+ TGG G +STGG GG+STGG
Sbjct: 622 TGGL*TGGFSTGGLYTGGFSTGGLYIGGFSTGGLYTGGFSTGGLYTGGFSTGG 464
Score = 60.1 bits (144), Expect = 8e-10
Identities = 48/116 (41%), Positives = 63/116 (53%), Gaps = 13/116 (11%)
Frame = -1
Query: 38 SGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGG---GGYDSGYNKTA-YSTDE 93
+GG Y G+ T GG+ TG T +ST ++GG GG+ +G T +ST
Sbjct: 847 TGGLYTGGFSTGGL*T--GGFSTGGLYTGGFSTGGFSTGGL*AGGFSTGGLYTGGFSTGG 674
Query: 94 PSGGGYGTGGA-GGGYSTGG----GYGTGGGGGGEYSTGG-GGGGYSTGG---GGY 140
GG+ TGG GG+STGG G+ TGG G +STGG GG+STGG GG+
Sbjct: 673 L*IGGFSTGGLYTGGFSTGGL*TGGFSTGGLYTGGFSTGGLYIGGFSTGGLYTGGF 506
Score = 52.4 bits (124), Expect = 2e-07
Identities = 50/129 (38%), Positives = 61/129 (46%), Gaps = 20/129 (15%)
Frame = -1
Query: 38 SGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGG---GGYDSGYNKTA-YSTDE 93
+GG Y G+ T GG+ TG T +ST GG GG+ +G T +ST
Sbjct: 652 TGGLYTGGFSTGGL*T--GGFSTGGLYTGGFST-----GGLYIGGFSTGGLYTGGFSTGG 494
Query: 94 PSGGGYGTGGA------GGGYSTG----GGYGTGGGGGGEYSTGG---GG---GGYSTGG 137
GG+ TGG GG+STG GG+ TGG G STGG GG GG STGG
Sbjct: 493 LYTGGFSTGGL*TGGW*TGGFSTGGL*IGGFSTGGL*TGGCSTGGLYTGGL*IGGCSTGG 314
Query: 138 GGYGNDDSG 146
G +G
Sbjct: 313 LYTGGISTG 287
Score = 51.2 bits (121), Expect = 4e-07
Identities = 48/117 (41%), Positives = 57/117 (48%), Gaps = 12/117 (10%)
Frame = -1
Query: 38 SGGDYDSGYKKSSYETSG---GGYETGYNKTSSYSTDEPNSGG---GGY-DSGYNKTAYS 90
+GG G + T G GG+ TG T +ST +GG GG G+ +S
Sbjct: 607 TGGFSTGGLYTGGFSTGGLYIGGFSTGGLYTGGFSTGGLYTGGFSTGGL*TGGW*TGGFS 428
Query: 91 TDEPSGGGYGTGGA-GGGYSTGGGYGTGGGGGGEYSTGG-GGGGYSTGG---GGYGN 142
T GG+ TGG GG STGG Y TGG G STGG GG STGG GG+ N
Sbjct: 427 TGGL*IGGFSTGGL*TGGCSTGGLY-TGGL*IGGCSTGGLYTGGISTGGW*IGGFSN 260
Score = 49.3 bits (116), Expect = 1e-06
Identities = 47/127 (37%), Positives = 60/127 (47%), Gaps = 18/127 (14%)
Frame = -1
Query: 32 YSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGG---GGYDSG----- 83
Y+ +GG Y G+ T GG+ TG T +ST +GG GG+ +G
Sbjct: 580 YTGGFSTGGLYIGGFSTGGLYT--GGFSTGGLYTGGFSTGGL*TGGW*TGGFSTGGL*IG 407
Query: 84 -YNKTAYSTDEPSGGGYGTGGAG-GGYSTGG----GYGTGGGGGGEYSTGG-GGGGYSTG 136
++ T S GG TGG GG STGG G TGG G +S GG GG+STG
Sbjct: 406 GFSTGGL*TGGCSTGGLYTGGL*IGGCSTGGLYTGGISTGGW*IGGFSNGGLYTGGFSTG 227
Query: 137 G---GGY 140
G GG+
Sbjct: 226 GL*IGGF 206
Score = 48.9 bits (115), Expect = 2e-06
Identities = 49/135 (36%), Positives = 60/135 (44%), Gaps = 26/135 (19%)
Frame = -1
Query: 32 YSTDEPSGGDYDSGYKKSSYETSG---GGYETGYNKTSSYSTDEPNSGG----GGYDSGY 84
Y+ +GG Y G+ T G GG+ TG +ST +GG G Y G
Sbjct: 520 YTGGFSTGGLYTGGFSTGGL*TGGW*TGGFSTGGL*IGGFSTGGL*TGGCSTGGLYTGGL 341
Query: 85 NKTAYSTDEPSGGGYGTGGAG-GGYSTGG----GYGTGGGGGGEYSTGG---GG------ 130
ST GG TGG GG+S GG G+ TGG G +STGG GG
Sbjct: 340 *IGGCSTGGLYTGGISTGGW*IGGFSNGGLYTGGFSTGGL*IGGFSTGGL*IGGCSIGGL 161
Query: 131 --GGYSTGG---GGY 140
GG+STGG GG+
Sbjct: 160 YTGGFSTGGL*NGGF 116
Score = 47.8 bits (112), Expect = 4e-06
Identities = 43/120 (35%), Positives = 55/120 (45%), Gaps = 17/120 (14%)
Frame = -1
Query: 38 SGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGG---GGYDSGYNKTAYSTDEP 94
+G Y G+ GG TG T T ++GG GG+ +G T
Sbjct: 907 TGXXYTXGF-------XXGGL*TGXXSTGGLYTGGFSTGGL*TGGFSTGGLYT------- 770
Query: 95 SGGGYGTGGA------GGGYSTG----GGYGTGGGGGGEYSTGG-GGGGYSTGG---GGY 140
GG+ TGG GG+STG GG+ TGG G +STGG GG+STGG GG+
Sbjct: 769 --GGFSTGGFSTGGL*AGGFSTGGLYTGGFSTGGL*IGGFSTGGLYTGGFSTGGL*TGGF 596
Score = 46.2 bits (108), Expect = 1e-05
Identities = 46/135 (34%), Positives = 58/135 (42%), Gaps = 26/135 (19%)
Frame = -1
Query: 32 YSTDEPSGGDYDSGYKKSSYETSG---GGYETGYNKTSSYSTDEPNSGG---------GG 79
Y+ +GG G+ + T G GG+ TG T ST +GG G
Sbjct: 490 YTGGFSTGGL*TGGW*TGGFSTGGL*IGGFSTGGL*TGGCSTGGLYTGGL*IGGCSTGGL 311
Query: 80 YDSGYNKTAYSTDEPSGGGYGTGGAG------GGYSTG----GGYGTGGGGGGEYSTGG- 128
Y G + + S GG TGG GG+STG GG GG G +STGG
Sbjct: 310 YTGGISTGGW*IGGFSNGGLYTGGFSTGGL*IGGFSTGGL*IGGCSIGGLYTGGFSTGGL 131
Query: 129 GGGGYSTGG---GGY 140
GG+STGG GG+
Sbjct: 130 *NGGFSTGG*YSGGF 86
>TC85601 nodulin
Length = 1933
Score = 61.2 bits (147), Expect = 3e-10
Identities = 50/121 (41%), Positives = 62/121 (50%), Gaps = 15/121 (12%)
Frame = -1
Query: 38 SGGDYDSGYKKSSYETSG---GGYETGYNKTSSYSTDEPNSGG---GGYDSGYNKTAYST 91
+GG G S+ T G GG+ TG T +ST SGG GG +G +ST
Sbjct: 634 TGGFSTGGL*TGSFSTGGLCTGGFSTGGLCTGGFSTRGL*SGGFSTGGLYTG----GFST 467
Query: 92 DEPSGGGYGTGG-AGGGYSTGG----GYGTGGGGGGEYSTGG-GGGGYSTGG---GGYGN 142
D G + TGG GG+STGG G+ TGG G +STGG GG+STGG GG+
Sbjct: 466 DGL*TGSFSTGGLCTGGFSTGGLCTGGFSTGGLCTGGFSTGGLCTGGFSTGGLCTGGFST 287
Query: 143 D 143
D
Sbjct: 286 D 284
Score = 54.7 bits (130), Expect = 3e-08
Identities = 50/131 (38%), Positives = 63/131 (47%), Gaps = 22/131 (16%)
Frame = -1
Query: 38 SGGDYDSGYKKSSYETSG---GGYETGYNKTSSYSTDEPNSGG---GGYDSGYNKTAYST 91
+GG G S+ T G GG+ TG T +ST +GG GG SG +ST
Sbjct: 1279 TGGFSTGGL*TGSFSTGGLCTGGFSTGGLCTGGFSTGGLCTGGFSTGGL*SG----GFST 1112
Query: 92 DEPSGGGYGTGGA-GGGYSTGG----GYGTGGGGGGEYSTGG---GG--------GGYST 135
GG+ TGG G +STGG G+ TGG G +STGG GG GG+ST
Sbjct: 1111 CGLYTGGFSTGGL*TGSFSTGGLCTGGFSTGGLCTGGFSTGGL*SGGFSTCGLYTGGFST 932
Query: 136 GGGGYGNDDSG 146
GG G+ +G
Sbjct: 931 GGL*TGSFSTG 899
Score = 54.3 bits (129), Expect = 4e-08
Identities = 47/121 (38%), Positives = 59/121 (47%), Gaps = 15/121 (12%)
Frame = -1
Query: 38 SGGDYDSGYKKSSYETSG---GGYETGYNKTSSYSTDEPNSGG---GGYDSGYNKTAYST 91
SGG SG + T G G + TG T +ST +GG GG +G +ST
Sbjct: 814 SGGFSTSGLYTGGFSTCGL*TGSFSTGGLCTGGFSTGGLCTGGFSTGGLFTG----GFST 647
Query: 92 DEPSGGGYGTGGA-GGGYSTGG----GYGTGGGGGGEYSTGG-GGGGYSTGG---GGYGN 142
GG+ TGG G +STGG G+ TGG G +ST G GG+STGG GG+
Sbjct: 646 CGLYTGGFSTGGL*TGSFSTGGLCTGGFSTGGLCTGGFSTRGL*SGGFSTGGLYTGGFST 467
Query: 143 D 143
D
Sbjct: 466 D 464
Score = 53.9 bits (128), Expect = 5e-08
Identities = 48/128 (37%), Positives = 61/128 (47%), Gaps = 25/128 (19%)
Frame = -1
Query: 38 SGGDYDSGYKKSSYETSG---GGYETGYNKTSSYSTDEPNSGG---GGYDSGYNKTAYST 91
+GG G + T G GG+ TG T +STD +G GG +G +ST
Sbjct: 574 TGGFSTGGLCTGGFSTRGL*SGGFSTGGLYTGGFSTDGL*TGSFSTGGLCTG----GFST 407
Query: 92 DEPSGGGYGTGG-AGGGYSTGG----GYGTGGGGGGEYSTGG--------GG---GGYST 135
GG+ TGG GG+STGG G+ TGG G +ST G GG GG+ST
Sbjct: 406 GGLCTGGFSTGGLCTGGFSTGGLCTGGFSTGGLCTGGFSTDGL*TGWCSTGGLYAGGFST 227
Query: 136 GG---GGY 140
GG GG+
Sbjct: 226 GGL*MGGF 203
Score = 53.5 bits (127), Expect = 7e-08
Identities = 48/134 (35%), Positives = 63/134 (46%), Gaps = 31/134 (23%)
Frame = -1
Query: 38 SGGDYDSGYKKSSYETSG---GGYETGYNKTSSYSTDEPNSGG--------GGYDSGYNK 86
+GG G S+ T G GG+ TG T +ST +GG GG+ +G
Sbjct: 784 TGGFSTCGL*TGSFSTGGLCTGGFSTGGLCTGGFSTGGLFTGGFSTCGLYTGGFSTGGL* 605
Query: 87 T-AYSTDEPSGGGYGTGGA-----------GGGYSTGG----GYGTGGGGGGEYSTGG-G 129
T ++ST GG+ TGG GG+STGG G+ T G G +STGG
Sbjct: 604 TGSFSTGGLCTGGFSTGGLCTGGFSTRGL*SGGFSTGGLYTGGFSTDGL*TGSFSTGGLC 425
Query: 130 GGGYSTGG---GGY 140
GG+STGG GG+
Sbjct: 424 TGGFSTGGLCTGGF 383
Score = 53.5 bits (127), Expect = 7e-08
Identities = 48/133 (36%), Positives = 61/133 (45%), Gaps = 33/133 (24%)
Frame = -1
Query: 38 SGGDYDSGYKKSSYETSG---GGYETGYNKTSSYSTDEPNSGG--------GGYDSGYNK 86
+G G + T G GG+ TG T +ST SGG GG+ +G
Sbjct: 1249 TGSFSTGGLCTGGFSTGGLCTGGFSTGGLCTGGFSTGGL*SGGFSTCGLYTGGFSTGGL* 1070
Query: 87 T-AYSTDEPSGGGYGTGG-AGGGYSTG--------------GGYGTGGGGGGEYSTGG-- 128
T ++ST GG+ TGG GG+STG GG+ TGG G +STGG
Sbjct: 1069 TGSFSTGGLCTGGFSTGGLCTGGFSTGGL*SGGFSTCGLYTGGFSTGGL*TGSFSTGGFS 890
Query: 129 -GG---GGYSTGG 137
GG GG+STGG
Sbjct: 889 TGGLCTGGFSTGG 851
Score = 52.8 bits (125), Expect = 1e-07
Identities = 45/115 (39%), Positives = 59/115 (51%), Gaps = 12/115 (10%)
Frame = -1
Query: 38 SGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGG---GGYDSGYNKTAYSTDEP 94
+GG + G+ T GG+ TG T S+ST +GG GG +G +ST
Sbjct: 679 TGGLFTGGFSTCGLYT--GGFSTGGL*TGSFSTGGLCTGGFSTGGLCTG----GFSTRGL 518
Query: 95 SGGGYGTGGA-GGGYSTGG----GYGTGGGGGGEYSTGG-GGGGYSTGG---GGY 140
GG+ TGG GG+ST G + TGG G +STGG GG+STGG GG+
Sbjct: 517 *SGGFSTGGLYTGGFSTDGL*TGSFSTGGLCTGGFSTGGLCTGGFSTGGLCTGGF 353
Score = 52.8 bits (125), Expect = 1e-07
Identities = 47/129 (36%), Positives = 62/129 (47%), Gaps = 26/129 (20%)
Frame = -1
Query: 38 SGGDYDSGYKKSSYETSG---GGYETGYNKTSSYSTDEPNSGG--------GGYDSG-YN 85
+GG G + T G GG+ TG T S+ST +GG GG+ +G
Sbjct: 1159 TGGFSTGGL*SGGFSTCGLYTGGFSTGGL*TGSFSTGGLCTGGFSTGGLCTGGFSTGGL* 980
Query: 86 KTAYSTDEPSGGGYGTGG------AGGGYSTG----GGYGTGGGGGGEYSTGG-GGGGYS 134
+ST GG+ TGG + GG+STG GG+ TGG G +STGG GG+S
Sbjct: 979 SGGFSTCGLYTGGFSTGGL*TGSFSTGGFSTGGLCTGGFSTGGLCIGVFSTGGL*SGGFS 800
Query: 135 TGG---GGY 140
T G GG+
Sbjct: 799 TSGLYTGGF 773
Score = 52.4 bits (124), Expect = 2e-07
Identities = 44/119 (36%), Positives = 54/119 (44%), Gaps = 11/119 (9%)
Frame = -1
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
GG Y G+ T GG+ TG T +ST GG Y G++ T S GG
Sbjct: 1381 GGLYSVGFST*GL*T--GGFSTGGL*TGGFST------GGLYTGGFSTGGL*TGSFSTGG 1226
Query: 99 YGTGGAGGGYSTGGGYGTGGGGGGEYSTGG---GG--------GGYSTGGGGYGNDDSG 146
TGG G GG+ TGG G +STGG GG GG+STGG G+ +G
Sbjct: 1225 LCTGGFSTGGLCTGGFSTGGLCTGGFSTGGL*SGGFSTCGLYTGGFSTGGL*TGSFSTG 1049
Score = 51.6 bits (122), Expect = 3e-07
Identities = 45/120 (37%), Positives = 56/120 (46%), Gaps = 17/120 (14%)
Frame = -1
Query: 38 SGGDYDSGYKKSSYETSG---GGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEP 94
+GG G + T G GG+ TG T S+ST GG G++ T
Sbjct: 1339 TGGFSTGGL*TGGFSTGGLYTGGFSTGGL*TGSFST------GGLCTGGFSTGGLCTGGF 1178
Query: 95 SGGGYGTGGA------GGGYST----GGGYGTGGGGGGEYSTGG-GGGGYSTGG---GGY 140
S GG TGG GG+ST GG+ TGG G +STGG GG+STGG GG+
Sbjct: 1177 STGGLCTGGFSTGGL*SGGFSTCGLYTGGFSTGGL*TGSFSTGGLCTGGFSTGGLCTGGF 998
Score = 50.1 bits (118), Expect = 8e-07
Identities = 42/123 (34%), Positives = 55/123 (44%), Gaps = 8/123 (6%)
Frame = -1
Query: 32 YSTDEPSGGDYDSGYKKSSYETSG---GGYETGYNKTSSYSTDEPNSGG----GGYDSGY 84
Y+ +GG + + T G GG+ TG +ST SGG G Y G+
Sbjct: 952 YTGGFSTGGL*TGSFSTGGFSTGGLCTGGFSTGGLCIGVFSTGGL*SGGFSTSGLYTGGF 773
Query: 85 NKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGG-GGGGYSTGGGGYGND 143
+ T S GG TGG G GG+ TGG G +ST G GG+STGG G+
Sbjct: 772 STCGL*TGSFSTGGLCTGGFSTGGLCTGGFSTGGLFTGGFSTCGLYTGGFSTGGL*TGSF 593
Query: 144 DSG 146
+G
Sbjct: 592 STG 584
Score = 47.0 bits (110), Expect = 7e-06
Identities = 44/118 (37%), Positives = 57/118 (48%), Gaps = 15/118 (12%)
Frame = -1
Query: 38 SGGDYDSGYKKSSYETSG---GGYETGYNKTSSYSTDEPNSGG---GGYDSGYNKTAYST 91
+GG G S+ T G GG+ TG T +ST +GG GG +G +ST
Sbjct: 484 TGGFSTDGL*TGSFSTGGLCTGGFSTGGLCTGGFSTGGLCTGGFSTGGLCTG----GFST 317
Query: 92 DEPSGGGYGTGGAGGGY-STGG----GYGTGGGGGGEYSTGGGG-GGYSTGG---GGY 140
GG+ T G G+ STGG G+ TGG G +S GG GG+ST G GG+
Sbjct: 316 GGLCTGGFSTDGL*TGWCSTGGLYAGGFSTGGL*MGGFSIGGWWVGGFSTSGLCTGGF 143
Score = 42.4 bits (98), Expect = 2e-04
Identities = 34/92 (36%), Positives = 41/92 (43%), Gaps = 1/92 (1%)
Frame = -1
Query: 56 GGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYG 115
GG+ G T +ST GG Y G++ T S GG TGG G G +
Sbjct: 1636 GGFSNGGL*TGGFST------GGLYTGGFSTGGLCTGGFSTGGLYTGGFSAGGL*TGSFS 1475
Query: 116 TGGGGGGEYSTGG-GGGGYSTGGGGYGNDDSG 146
TG G +STGG G+STGG GN G
Sbjct: 1474 TGDLCTGGFSTGGLCT*GFSTGGL*TGNFSIG 1379
Score = 38.5 bits (88), Expect = 0.002
Identities = 40/119 (33%), Positives = 51/119 (42%), Gaps = 16/119 (13%)
Frame = -1
Query: 38 SGGDYDSGYKKSSYETSG---GGYETGYNKTSSYSTDEPNSG----GGGYDSGYNKTAYS 90
+GG G + T G GG+ TG T +STD +G GG Y
Sbjct: 394 TGGFSTGGLCTGGFSTGGLCTGGFSTGGLCTGGFSTDGL*TGWCSTGGLY---------- 245
Query: 91 TDEPSGGGYGTGGA-GGGYSTG----GGYGTGGGGGGEYSTGG-GGGGYSTGG---GGY 140
GG+ TGG GG+S G GG+ T G G +S G G+STGG GG+
Sbjct: 244 -----AGGFSTGGL*MGGFSIGGWWVGGFSTSGLCTGGFSMCGL*NWGFSTGGLYSGGF 83
Score = 37.4 bits (85), Expect = 0.005
Identities = 28/58 (48%), Positives = 33/58 (56%), Gaps = 9/58 (15%)
Frame = -1
Query: 92 DEPSGGGYGTGGAG-GGYSTGG----GYGTGGGGGGEYSTGG-GGGGYSTGG---GGY 140
D S GG GG GG+S GG G+ TGG G +STGG GG+STGG GG+
Sbjct: 1681 DFGS*GGI*NGGV*IGGFSNGGL*TGGFSTGGLYTGGFSTGGLCTGGFSTGGLYTGGF 1508
Score = 37.0 bits (84), Expect = 0.007
Identities = 26/55 (47%), Positives = 33/55 (59%), Gaps = 2/55 (3%)
Frame = -1
Query: 95 SGGGYGTGG-AGGGYSTGGGYGTGGGGGGEYSTGG-GGGGYSTGGGGYGNDDSGN 147
S GG TGG + GG T GG+ TGG G +STGG GG+S GG G+ +G+
Sbjct: 1627 SNGGL*TGGFSTGGLYT-GGFSTGGLCTGGFSTGGLYTGGFSAGGL*TGSFSTGD 1466
Score = 32.0 bits (71), Expect = 0.22
Identities = 24/55 (43%), Positives = 29/55 (52%), Gaps = 7/55 (12%)
Frame = -1
Query: 93 EPSGGGYGTGGAGGGYSTG---GGYGTGGGGGGEYSTGG-GGGGYSTGG---GGY 140
EPS G GG + G GG+ GG G +STGG GG+STGG GG+
Sbjct: 1702 EPSSAREDFGS*GGI*NGGV*IGGFSNGGL*TGGFSTGGLYTGGFSTGGLCTGGF 1538
>BM814143 GP|21322711|e pherophorin-dz1 protein {Volvox carteri f.
nagariensis}, partial (4%)
Length = 132
Score = 61.2 bits (147), Expect = 3e-10
Identities = 29/44 (65%), Positives = 29/44 (65%)
Frame = +1
Query: 96 GGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGG 139
GGG G GG GGG GGG G GGGGGG GGGGGG GGGG
Sbjct: 1 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 132
Score = 59.3 bits (142), Expect = 1e-09
Identities = 30/46 (65%), Positives = 30/46 (65%)
Frame = +3
Query: 96 GGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYG 141
GGG G GG GGG GGG G GGGGGG GGGGGG GGGG G
Sbjct: 6 GGGGGGGGGGGGGGGGGGGGGGGGGGG----GGGGGGGGGGGGGGG 131
Score = 59.3 bits (142), Expect = 1e-09
Identities = 30/46 (65%), Positives = 30/46 (65%)
Frame = +2
Query: 96 GGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYG 141
GGG G GG GGG GGG G GGGGGG GGGGGG GGGG G
Sbjct: 2 GGGGGGGGGGGGGGGGGGGGGGGGGGG----GGGGGGGGGGGGGGG 127
Score = 58.9 bits (141), Expect = 2e-09
Identities = 30/46 (65%), Positives = 30/46 (65%)
Frame = +1
Query: 96 GGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYG 141
GGG G GG GGG GGG G GGGGGG GGGGGG GGGG G
Sbjct: 4 GGGGGGGGGGGG---GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 132
Score = 53.1 bits (126), Expect = 9e-08
Identities = 27/45 (60%), Positives = 27/45 (60%)
Frame = +2
Query: 102 GGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSG 146
GG GGG GGG G GGGGGG GGGGGG GGGG G G
Sbjct: 5 GGGGGG---GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 130
Score = 53.1 bits (126), Expect = 9e-08
Identities = 27/45 (60%), Positives = 27/45 (60%)
Frame = +3
Query: 102 GGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSG 146
GG GGG GGG G GGGGGG GGGGGG GGGG G G
Sbjct: 3 GGGGGG---GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 128
>TC84888 similar to GP|21322752|dbj|BAB78536. cold shock protein-1 {Triticum
aestivum}, partial (58%)
Length = 612
Score = 60.5 bits (145), Expect = 6e-10
Identities = 35/71 (49%), Positives = 37/71 (51%), Gaps = 5/71 (7%)
Frame = +2
Query: 76 GGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTG-GGGGGYS 134
GGGGY GY++ GYG GG G GGG G G GGGG G GGGGGY
Sbjct: 323 GGGGYGGGYDQ-----------GYGGGGGSYGGGFGGGRGGGRGGGGYXQGGYGGGGGYX 469
Query: 135 TGG----GGYG 141
GG GGYG
Sbjct: 470 QGGYGGQGGYG 502
Score = 58.5 bits (140), Expect = 2e-09
Identities = 46/139 (33%), Positives = 58/139 (41%), Gaps = 10/139 (7%)
Frame = +2
Query: 18 DRPTDTETGYNKTS-YSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDE--PN 74
++ T T +N T + P GG D ++S +T G + + E
Sbjct: 53 EKQTGTVKWFNSTKGFGFISPDGGGEDLFVHQTSIKTDG------FRSLAEGEAVEFIVE 214
Query: 75 SGGGGYDSGYNKTAYSTDEPSGG-----GYGTGGAGGGYSTGGGYGTG-GGGGGEYSTG- 127
+G G + T + P G GYG GG GG GGGY G GGGGG Y G
Sbjct: 215 AGDDGRTKAIDVTGPNGAPPQGAPRQEMGYGGGGGRGGGGYGGGYDQGYGGGGGSYGGGF 394
Query: 128 GGGGGYSTGGGGYGNDDSG 146
GGG G GGGGY G
Sbjct: 395 GGGRGGGRGGGGYXQGGYG 451
Score = 40.0 bits (92), Expect = 8e-04
Identities = 27/76 (35%), Positives = 29/76 (37%)
Frame = +2
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
GG Y GY + Y GG Y G+ GGG GY GGG
Sbjct: 326 GGGYGGGYDQG-YGGGGGSYGGGFGGGRGGG----RGGGGYXQGGYG---------GGGG 463
Query: 99 YGTGGAGGGYSTGGGY 114
Y GG GG GGGY
Sbjct: 464 YXQGGYGGQGGYGGGY 511
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.303 0.131 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,767,855
Number of Sequences: 36976
Number of extensions: 136084
Number of successful extensions: 12577
Number of sequences better than 10.0: 621
Number of HSP's better than 10.0 without gapping: 1861
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5071
length of query: 243
length of database: 9,014,727
effective HSP length: 93
effective length of query: 150
effective length of database: 5,575,959
effective search space: 836393850
effective search space used: 836393850
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 17 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.7 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0052b.4


