

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0051b.10
(187 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
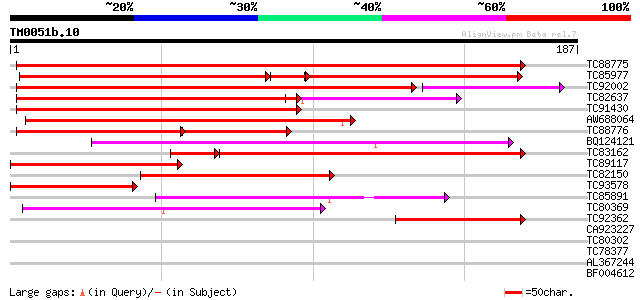
Score E
Sequences producing significant alignments: (bits) Value
TC88775 homologue to PIR|F86427|F86427 auxin response factor 6 (... 190 3e-49
TC85977 similar to GP|10176918|dbj|BAB10162. auxin response fact... 121 1e-41
TC92002 similar to GP|4104929|gb|AAD02218.1| auxin response fact... 146 4e-40
TC82637 similar to PIR|G86331|G86331 IAA24 [imported] - Arabidop... 125 1e-34
TC91430 similar to PIR|F86427|F86427 auxin response factor 6 (AR... 132 1e-31
AW688064 homologue to GP|13272405|gb unknown protein {Arabidopsi... 125 1e-29
TC88776 homologue to PIR|F86427|F86427 auxin response factor 6 (... 89 2e-28
BQ124121 similar to PIR|G86331|G863 IAA24 [imported] - Arabidops... 110 3e-25
TC83162 homologue to PIR|F86427|F86427 auxin response factor 6 (... 94 8e-23
TC89117 similar to GP|12484199|gb|AAG53998.1 auxin response tran... 99 7e-22
TC82150 similar to GP|19352051|dbj|BAB85919. auxin response fact... 91 3e-19
TC93578 similar to GP|9757747|dbj|BAB08228.1 auxin response fact... 77 3e-15
TC85891 similar to PIR|A86386|A86386 probable DNA-binding protei... 49 1e-06
TC80369 similar to PIR|A86386|A86386 probable DNA-binding protei... 45 2e-05
TC92362 similar to GP|19352047|dbj|BAB85917. auxin response fact... 42 2e-04
CA923227 similar to GP|20152530|emb putative auxin response fact... 35 0.013
TC80302 33 0.065
TC78377 similar to GP|16974560|gb|AAL31196.1 At2g02870/T17M13.4 ... 31 0.32
AL367244 similar to PIR|C86468|C864 probable auxin response fact... 31 0.32
BF004612 similar to PIR|G86331|G863 IAA24 [imported] - Arabidops... 30 0.55
>TC88775 homologue to PIR|F86427|F86427 auxin response factor 6 (ARF6)
[imported] - Arabidopsis thaliana, partial (23%)
Length = 678
Score = 190 bits (482), Expect = 3e-49
Identities = 89/168 (52%), Positives = 120/168 (70%)
Frame = +3
Query: 3 SHMFCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCHIY 62
++ FCKTLTA DT TH GFSVPRRAAE PPLD+ QQ P+QEL+A+DLHG EWKF HI+
Sbjct: 15 TNYFCKTLTASDTSTHGGFSVPRRAAEKVFPPLDFSQQPPAQELIARDLHGNEWKFRHIF 194
Query: 63 RG*PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLGIRRSVGPRNGLPESTVGNQ 122
RG P+ HLLTTGWS+FVS LV+GD+V F+ E +L LGIRR+ P+ +P S + +
Sbjct: 195 RGQPKRHLLTTGWSVFVSAKRLVAGDSVLFIWNEKNQLLLGIRRASRPQTVMPSSVLSSD 374
Query: 123 NSYPHILSVVANVISTRSMFHVFYSPRASNSDFVFPYQKYAKSIHDNK 170
+ + +L+ A+ +T S F +FY+PRA S+FV P KY K+++ +
Sbjct: 375 SMHLGLLAAAAHAAATNSRFTIFYNPRACPSEFVIPLAKYVKAVYHTR 518
>TC85977 similar to GP|10176918|dbj|BAB10162. auxin response factor-like
protein {Arabidopsis thaliana}, partial (65%)
Length = 3162
Score = 121 bits (304), Expect(3) = 1e-41
Identities = 56/83 (67%), Positives = 66/83 (79%)
Frame = +3
Query: 4 HMFCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCHIYR 63
H FCKTLTA DT TH GFSV +R A++CLPPLD +Q P+QELVAKDLHG EW+F HI+R
Sbjct: 609 HSFCKTLTASDTSTHGGFSVLKRHADECLPPLDMSKQPPTQELVAKDLHGNEWRFRHIFR 788
Query: 64 G*PRWHLLTTGWSIFVSQNNLVS 86
G PR HLL +GWS+FVS LV+
Sbjct: 789 GQPRRHLLQSGWSVFVSSKRLVA 857
Score = 58.9 bits (141), Expect(3) = 1e-41
Identities = 23/72 (31%), Positives = 46/72 (62%)
Frame = +2
Query: 98 GELRLGIRRSVGPRNGLPESTVGNQNSYPHILSVVANVISTRSMFHVFYSPRASNSDFVF 157
G LR+G+RR++ + +P S + + + + +L+ + + T +MF V+Y PR S ++F+
Sbjct: 893 GSLRVGVRRAMRQQGNVPSSVISSHSMHLGVLATAWHAVLTGTMFTVYYKPRTSPAEFIV 1072
Query: 158 PYQKYAKSIHDN 169
PY +Y +S+ +N
Sbjct: 1073PYDQYMESLKNN 1108
Score = 26.2 bits (56), Expect(3) = 1e-41
Identities = 11/13 (84%), Positives = 11/13 (84%)
Frame = +1
Query: 87 GDAVSFLRGENGE 99
GDA FLRGENGE
Sbjct: 859 GDAFIFLRGENGE 897
>TC92002 similar to GP|4104929|gb|AAD02218.1| auxin response factor 7
{Arabidopsis thaliana}, partial (31%)
Length = 1138
Score = 146 bits (368), Expect(2) = 4e-40
Identities = 72/132 (54%), Positives = 90/132 (67%)
Frame = +3
Query: 3 SHMFCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCHIY 62
S FCKTLTA DT TH GFSVPRRAAE PPLD+ Q P+QE+VAKDL+ W F HIY
Sbjct: 186 SEFFCKTLTASDTSTHGGFSVPRRAAEKIFPPLDFSMQPPAQEIVAKDLYDTTWTFRHIY 365
Query: 63 RG*PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLGIRRSVGPRNGLPESTVGNQ 122
RG P+ HLLTTGWS+FVS L +GD+V F+R E +L LGI+RS + L S + +
Sbjct: 366 RGQPKRHLLTTGWSVFVSTKRLFAGDSVLFIRDEKQQLLLGIKRSSRQQPALSSSVISSD 545
Query: 123 NSYPHILSVVAN 134
+ + IL+ A+
Sbjct: 546 SMHIGILAAAAH 581
Score = 35.0 bits (79), Expect(2) = 4e-40
Identities = 17/47 (36%), Positives = 26/47 (55%)
Frame = +1
Query: 137 STRSMFHVFYSPRASNSDFVFPYQKYAKSIHDNKRNEPSHRVMEERD 183
S S F +FY+PR S +FV P KY K+++ + R+M E +
Sbjct: 589 SNNSPFTIFYNPRTSPCEFVIPLAKYNKALYTHVSLGMRFRMMFETE 729
>TC82637 similar to PIR|G86331|G86331 IAA24 [imported] - Arabidopsis
thaliana, partial (28%)
Length = 889
Score = 125 bits (314), Expect(2) = 1e-34
Identities = 60/94 (63%), Positives = 68/94 (71%)
Frame = +1
Query: 3 SHMFCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCHIY 62
+ FCKTLTA DT TH GFSVPRRAAE PPLDY Q P+QELV +DLH W F HIY
Sbjct: 439 TEFFCKTLTASDTSTHGGFSVPRRAAEKLFPPLDYTIQPPTQELVVRDLHDNTWTFRHIY 618
Query: 63 RG*PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGE 96
RG P+ HLLTTGWS+FV L +GD+V F+R E
Sbjct: 619 RGQPKRHLLTTGWSLFVGSKRLRAGDSVLFIRDE 720
Score = 37.4 bits (85), Expect(2) = 1e-34
Identities = 19/59 (32%), Positives = 33/59 (55%), Gaps = 1/59 (1%)
Frame = +2
Query: 92 FLRG-ENGELRLGIRRSVGPRNGLPESTVGNQNSYPHILSVVANVISTRSMFHVFYSPR 149
FL G + +LR+G+RR+ + LP S + + +L+ A+ + R F +FY+PR
Sbjct: 704 FLSGMKKSQLRVGVRRANRQQTTLPSSVLSADSCTFGVLAAAAHAAANRKSFTIFYNPR 880
>TC91430 similar to PIR|F86427|F86427 auxin response factor 6 (ARF6)
[imported] - Arabidopsis thaliana, partial (23%)
Length = 728
Score = 132 bits (331), Expect = 1e-31
Identities = 61/94 (64%), Positives = 74/94 (77%)
Frame = +3
Query: 3 SHMFCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCHIY 62
++ FCK LTA DT TH GFSVPRRAAE PPLD+ QQ P+QEL+A+DLHG +WKF HI+
Sbjct: 429 TNYFCKILTASDTSTHGGFSVPRRAAEKVFPPLDFTQQPPAQELIARDLHGNDWKFKHIF 608
Query: 63 RG*PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGE 96
RG P+ HLLTTGWS+FVS LV+GD+V F+ E
Sbjct: 609 RGQPKRHLLTTGWSVFVSAKRLVAGDSVLFIWNE 710
>AW688064 homologue to GP|13272405|gb unknown protein {Arabidopsis thaliana},
partial (15%)
Length = 576
Score = 125 bits (313), Expect = 1e-29
Identities = 65/110 (59%), Positives = 76/110 (69%), Gaps = 1/110 (0%)
Frame = +1
Query: 6 FCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCHIYRG* 65
F KTLT D GFSVPR AE P LDY + P Q ++AKD+HG WKF HIYRG
Sbjct: 97 FAKTLTQSDANNGGGFSVPRYCAETIFPRLDYSAEPPVQTVIAKDVHGEVWKFRHIYRGT 276
Query: 66 PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLGIRRSV-GPRNGL 114
PR HLLTTGWS FV+Q LV+GD++ FLR E+GEL +GIRR+ G NGL
Sbjct: 277 PRRHLLTTGWSSFVNQKKLVAGDSIVFLRAESGELFVGIRRAKRGIVNGL 426
>TC88776 homologue to PIR|F86427|F86427 auxin response factor 6 (ARF6)
[imported] - Arabidopsis thaliana, partial (22%)
Length = 1278
Score = 89.0 bits (219), Expect(2) = 2e-28
Identities = 40/56 (71%), Positives = 46/56 (81%)
Frame = +1
Query: 3 SHMFCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKF 58
++ FCKTLTA DT TH GFSVPRRAAE PPLD+ QQ P+QEL+A+DLHG EWKF
Sbjct: 886 TNYFCKTLTASDTSTHGGFSVPRRAAEKVFPPLDFSQQPPAQELIARDLHGNEWKF 1053
Score = 53.1 bits (126), Expect(2) = 2e-28
Identities = 23/36 (63%), Positives = 29/36 (79%)
Frame = +2
Query: 58 FCHIYRG*PRWHLLTTGWSIFVSQNNLVSGDAVSFL 93
F HI+RG P+ HLLTTGWS+FVS LV+GD+V F+
Sbjct: 1052 FRHIFRGQPKRHLLTTGWSVFVSAKRLVAGDSVLFI 1159
>BQ124121 similar to PIR|G86331|G863 IAA24 [imported] - Arabidopsis thaliana,
partial (2%)
Length = 569
Score = 110 bits (275), Expect = 3e-25
Identities = 58/141 (41%), Positives = 82/141 (58%), Gaps = 2/141 (1%)
Frame = +2
Query: 28 AEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCHIYRG*PRWHLLTTGWSIFVSQNNLVSG 87
A+ C P LD Q P Q+LV KDL G+EW F HIY R HLLT GW+ FV+ NL G
Sbjct: 5 ADRCFP*LDMTLQPPMQDLVVKDLQGIEWNFRHIYCDHERAHLLTNGWNTFVNSKNLRPG 184
Query: 88 DAVSFLRGENGELRLGIRRSVGPRNGLPESTV--GNQNSYPHILSVVANVISTRSMFHVF 145
D+ F+ GENGE+ +GIRR++ + + +QN L+ V + +S S+FH+
Sbjct: 185 DSCIFVSGENGEIGIGIRRAMKQHSHICTKLCQQSSQNIQLGALAAVVHAVSIGSLFHLQ 364
Query: 146 YSPRASNSDFVFPYQKYAKSI 166
Y P + +F+ P + Y +SI
Sbjct: 365 YHPWIAPFEFMIPLKTYVESI 427
>TC83162 homologue to PIR|F86427|F86427 auxin response factor 6 (ARF6)
[imported] - Arabidopsis thaliana, partial (18%)
Length = 569
Score = 94.0 bits (232), Expect(2) = 8e-23
Identities = 44/101 (43%), Positives = 67/101 (65%)
Frame = +2
Query: 70 LLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLGIRRSVGPRNGLPESTVGNQNSYPHIL 129
LLTTGWS+FVS LV+GD+V F+ E +L LGIRR+ P+ +P S + + + + +L
Sbjct: 62 LLTTGWSVFVSAKRLVAGDSVLFIWNEKNQLLLGIRRASRPQTVMPSSVLSSDSMHIGLL 241
Query: 130 SVVANVISTRSMFHVFYSPRASNSDFVFPYQKYAKSIHDNK 170
+ A+ +T S F VF++PRAS S+FV P KY K+++ +
Sbjct: 242 AAAAHAAATNSCFTVFFNPRASPSEFVIPLSKYIKAVYHTR 364
Score = 29.3 bits (64), Expect(2) = 8e-23
Identities = 11/16 (68%), Positives = 13/16 (80%)
Frame = +1
Query: 54 VEWKFCHIYRG*PRWH 69
VEWKF HI+RG P+ H
Sbjct: 13 VEWKFRHIFRGQPKRH 60
>TC89117 similar to GP|12484199|gb|AAG53998.1 auxin response transcription
factor 3 {Arabidopsis thaliana}, partial (26%)
Length = 861
Score = 99.4 bits (246), Expect = 7e-22
Identities = 45/57 (78%), Positives = 48/57 (83%)
Frame = +1
Query: 1 STSHMFCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWK 57
+T HMFCKTLTA DT TH GFSVPR AAEDC PPLDY QQRPSQELV KDLHG +W+
Sbjct: 670 TTPHMFCKTLTASDTSTHGGFSVPRXAAEDCFPPLDYGQQRPSQELVXKDLHGSKWE 840
>TC82150 similar to GP|19352051|dbj|BAB85919. auxin response factor 10
{Oryza sativa}, partial (30%)
Length = 1263
Score = 90.5 bits (223), Expect = 3e-19
Identities = 41/64 (64%), Positives = 51/64 (79%)
Frame = +1
Query: 44 QELVAKDLHGVEWKFCHIYRG*PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLG 103
Q ++AKD+HG WKF HIYRG PR HLLTTGWS FV+ LV+GD++ FLR ENG+L +G
Sbjct: 1 QTIIAKDMHGQCWKFKHIYRGTPRRHLLTTGWSNFVNHKKLVAGDSIVFLRAENGDLCVG 180
Query: 104 IRRS 107
IRR+
Sbjct: 181 IRRA 192
>TC93578 similar to GP|9757747|dbj|BAB08228.1 auxin response factor 4
{Arabidopsis thaliana}, partial (18%)
Length = 634
Score = 77.4 bits (189), Expect = 3e-15
Identities = 35/42 (83%), Positives = 35/42 (83%)
Frame = +2
Query: 1 STSHMFCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRP 42
ST HMFCKTLT DT TH GFSVPRRAAEDC PPLDYK QRP
Sbjct: 506 STPHMFCKTLTVSDTSTHGGFSVPRRAAEDCFPPLDYKLQRP 631
>TC85891 similar to PIR|A86386|A86386 probable DNA-binding protein RAV2
[imported] - Arabidopsis thaliana, partial (57%)
Length = 2083
Score = 48.5 bits (114), Expect = 1e-06
Identities = 32/100 (32%), Positives = 50/100 (50%), Gaps = 3/100 (3%)
Frame = +2
Query: 49 KDLHGVEWKFCHIYRG*PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLGI---R 105
+D+ G W+F + Y + ++LT GWS FV + NL +GDAV F R + +L I
Sbjct: 1328 EDVGGKVWRFRYSYWNSSQSYVLTKGWSRFVKEKNLRAGDAVRFFRSTGPDRQLYIDCKA 1507
Query: 106 RSVGPRNGLPESTVGNQNSYPHILSVVANVISTRSMFHVF 145
RS+G G ++ N N+ + V V+ M +F
Sbjct: 1508 RSIGVVGGQVDN---NNNNTGGLFITVRPVVEPVQMVRLF 1618
>TC80369 similar to PIR|A86386|A86386 probable DNA-binding protein RAV2
[imported] - Arabidopsis thaliana, partial (30%)
Length = 821
Score = 44.7 bits (104), Expect = 2e-05
Identities = 32/107 (29%), Positives = 47/107 (43%), Gaps = 7/107 (6%)
Frame = +3
Query: 5 MFCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAK-------DLHGVEWK 57
+F K +T D +P++ AE P + S K D+ G W+
Sbjct: 15 LFEKVVTPSDVGKLNRLVIPKQHAEKHFPLQKADCVQGSASAAGKGVLLNFEDIGGKVWR 194
Query: 58 FCHIYRG*PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLGI 104
F + Y + ++LT GWS FV + NL +GD V F R E +L I
Sbjct: 195 FRYSYWNSSQSYVLTKGWSRFVKEKNLKAGDTVCFQRSTGPEKQLFI 335
>TC92362 similar to GP|19352047|dbj|BAB85917. auxin response factor 7b
{Oryza sativa}, partial (11%)
Length = 399
Score = 41.6 bits (96), Expect = 2e-04
Identities = 18/43 (41%), Positives = 29/43 (66%)
Frame = +2
Query: 128 ILSVVANVISTRSMFHVFYSPRASNSDFVFPYQKYAKSIHDNK 170
IL+ A+ + S F VFY+PRAS S+FV P KY ++++ ++
Sbjct: 155 ILAAAAHASANNSPFTVFYNPRASPSEFVIPLAKYYRAVYSHQ 283
>CA923227 similar to GP|20152530|emb putative auxin response factor 32
{Arabidopsis thaliana}, partial (25%)
Length = 238
Score = 35.4 bits (80), Expect = 0.013
Identities = 20/56 (35%), Positives = 28/56 (49%)
Frame = -3
Query: 49 KDLHGVEWKFCHIYRG*PRWHLLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLGI 104
+D G W+F + Y + ++LT GWS +V L +GD V F R RL I
Sbjct: 194 EDESGKCWRFRYFYWNSSQSYVLTKGWSRYVKDKRLDAGDVVLFERHRVDSQRLFI 27
>TC80302
Length = 886
Score = 33.1 bits (74), Expect = 0.065
Identities = 29/105 (27%), Positives = 44/105 (41%), Gaps = 1/105 (0%)
Frame = +3
Query: 1 STSHMFCKTLTAFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCH 60
ST F K + F+ R +VP + + LP K ++ ++L G +W
Sbjct: 291 STFPYFVKIIKTFNVDGPRILNVPHQFSIAHLPNGKIK-------IILRNLKGEQWTANS 449
Query: 61 IYRG*PRW-HLLTTGWSIFVSQNNLVSGDAVSFLRGENGELRLGI 104
+ R H L GW FV NN+ GD F + ELR+ +
Sbjct: 450 VPRSRVHTSHTLCGGWMSFVRANNIKLGDVCVFELINDCELRVRV 584
>TC78377 similar to GP|16974560|gb|AAL31196.1 At2g02870/T17M13.4
{Arabidopsis thaliana}, partial (75%)
Length = 1908
Score = 30.8 bits (68), Expect = 0.32
Identities = 23/71 (32%), Positives = 32/71 (44%)
Frame = +2
Query: 12 AFDTITHRGFSVPRRAAEDCLPPLDYKQQRPSQELVAKDLHGVEWKFCHIYRG*PRWHLL 71
A+D I R +PR A+ DC D + EL+ + G E + IY R+ LL
Sbjct: 878 AYDPIRQRWMHLPRMASNDCFMCSDKESLAVGTELL---VFGRELRSHVIY----RYSLL 1036
Query: 72 TTGWSIFVSQN 82
T WS + N
Sbjct: 1037TNSWSSGMRMN 1069
>AL367244 similar to PIR|C86468|C864 probable auxin response factor
53188-50111 [imported] - Arabidopsis thaliana, partial
(3%)
Length = 199
Score = 30.8 bits (68), Expect = 0.32
Identities = 13/24 (54%), Positives = 16/24 (66%)
Frame = +3
Query: 66 PRWHLLTTGWSIFVSQNNLVSGDA 89
P+ HLL TGWS F ++ LV DA
Sbjct: 60 PQRHLLITGWSAFSNKKKLVCSDA 131
>BF004612 similar to PIR|G86331|G863 IAA24 [imported] - Arabidopsis thaliana,
partial (8%)
Length = 507
Score = 30.0 bits (66), Expect = 0.55
Identities = 12/22 (54%), Positives = 15/22 (67%)
Frame = +3
Query: 146 YSPRASNSDFVFPYQKYAKSIH 167
Y+PRA SDFV P KY K ++
Sbjct: 3 YNPRACPSDFVIPLAKYRKCVY 68
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.137 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,685,154
Number of Sequences: 36976
Number of extensions: 94896
Number of successful extensions: 473
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 472
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 472
length of query: 187
length of database: 9,014,727
effective HSP length: 91
effective length of query: 96
effective length of database: 5,649,911
effective search space: 542391456
effective search space used: 542391456
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0051b.10


