

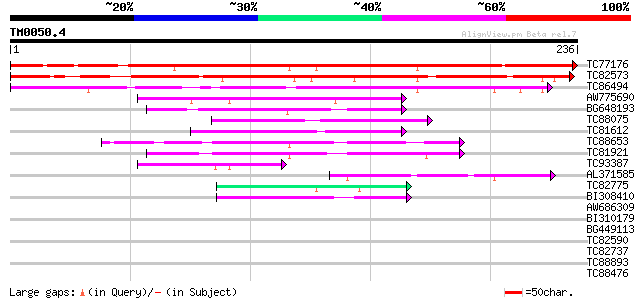
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0050.4
(236 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77176 homologue to GP|7228329|emb|CAB77055.1 putative TFIIIA (... 303 5e-83
TC82573 similar to PIR|T09602|T09602 probable zinc finger protei... 255 9e-69
TC86494 homologue to GP|7228329|emb|CAB77055.1 putative TFIIIA (... 158 2e-39
AW775690 similar to GP|8567776|gb|A hypothetical protein {Arabid... 77 4e-15
BG648193 similar to GP|2346976|dbj| ZPT2-13 {Petunia x hybrida},... 77 4e-15
TC88075 homologue to GP|2346978|dbj|BAA21923.1 ZPT2-14 {Petunia ... 75 3e-14
TC81612 similar to GP|2346978|dbj|BAA21923.1 ZPT2-14 {Petunia x ... 74 6e-14
TC88653 similar to GP|1786134|dbj|BAA19110.1 PEThy;ZPT2-5 {Petun... 72 1e-13
TC81921 homologue to GP|2346978|dbj|BAA21923.1 ZPT2-14 {Petunia ... 72 2e-13
TC93387 similar to GP|1786136|dbj|BAA19111.1 PEThy;ZPT2-6 {Petun... 54 5e-08
AL371585 similar to PIR|S69193|S691 probable finger protein Pszf... 50 6e-07
TC82775 similar to PIR|T52382|T52382 zinc finger protein ZPT4-4 ... 45 2e-05
BI308410 similar to PIR|S55882|S558 CCHH finger protein 2 - Arab... 41 5e-04
AW686309 homologue to PIR|S55886|S558 CCHH finger protein 6 - Ar... 38 0.004
BI310179 similar to PIR|T52381|T523 zinc finger protein Zat1 C2... 37 0.007
BG449113 similar to PIR|S55881|S558 CCHH finger protein 1 - Arab... 37 0.009
TC82590 similar to PIR|T50806|T50806 zinc finger-like protein - ... 37 0.009
TC82737 homologue to PIR|B96686|B96686 probable C2H2-type zinc f... 36 0.011
TC88893 similar to GP|6478938|gb|AAF14043.1| putative C2H2-type ... 36 0.015
TC88476 homologue to GP|21592423|gb|AAM64374.1 zinc finger prote... 36 0.015
>TC77176 homologue to GP|7228329|emb|CAB77055.1 putative TFIIIA (or
kruppel)-like zinc finger protein {Medicago sativa},
partial (96%)
Length = 1005
Score = 303 bits (775), Expect = 5e-83
Identities = 164/243 (67%), Positives = 185/243 (75%), Gaps = 7/243 (2%)
Frame = +2
Query: 1 MAMEALKSPTTAAPTFTPFQEPNHSYMVDAPWAKRKRSKRSRTDSHHNHASCTEEEYLAL 60
MAMEAL SPTTA P FTPF+EPN SY+ + PW K KRSKRSR D +SCTEEEYLAL
Sbjct: 110 MAMEALNSPTTATP-FTPFEEPNLSYL-ETPWTKGKRSKRSRMDQ----SSCTEEEYLAL 271
Query: 61 CLIMLAR-GSTAVTPKLTLSRPAPVTAEKLSYKCSVCEKTFPSYQALGGHKASHRK--LA 117
CLIMLAR G+ ++ PAP+T KLS+KCSVC K F SYQALGGHKASHRK ++
Sbjct: 272 CLIMLARSGNNNDNKTESVPVPAPLTTVKLSHKCSVCNKAFSSYQALGGHKASHRKAVMS 451
Query: 118 GAAAEDHST--SSAVTTSSASNGGGKVHQCSICQKSFPTGQALGGHKRCHYEG--GGGAS 173
ED +T SSAVTTSSASNG K H+CSIC KSFPTGQALGGHKRCHYEG G GA
Sbjct: 452 ATTVEDQTTTTSSAVTTSSASNGKNKTHECSICHKSFPTGQALGGHKRCHYEGSVGAGAG 631
Query: 174 STATATASEGVGSTHSHQRNFDLNLPAFPDFSASKFFVEEEVSSPLPSKKPRLLPKIEIP 233
S+A ASEGVGS+HSH R+FDLNLPAFPDFS KFFV++EVSSPLP+ K L K+EIP
Sbjct: 632 SSAVTAASEGVGSSHSHHRDFDLNLPAFPDFS-KKFFVDDEVSSPLPAAKKPCLFKLEIP 808
Query: 234 HYY 236
+Y
Sbjct: 809 SHY 817
>TC82573 similar to PIR|T09602|T09602 probable zinc finger protein SCOF-1
cold-inducible - soybean, partial (65%)
Length = 857
Score = 255 bits (652), Expect = 9e-69
Identities = 154/252 (61%), Positives = 174/252 (68%), Gaps = 17/252 (6%)
Frame = +3
Query: 1 MAMEALKSPTTAAPTFTPFQEPNHSYMVDAPWAKRKRSKRSRTDSHHNHASCTEEEYLAL 60
MA+EAL SPT+ P FT F EPN PW KRKRSKRSR SCTEEEYLAL
Sbjct: 39 MALEALNSPTSTTPKFT-FDEPN------LPWTKRKRSKRSR--------SCTEEEYLAL 173
Query: 61 CLIMLARGSTAVTPKLTLSRPAPVTAE------KLSYKCSVCEKTFPSYQALGGHKASHR 114
CLIMLARG T L+ P P T + KLSYKCSVC K F SYQALGGHKASHR
Sbjct: 174 CLIMLARGHTNRHDFNPLN-PPPTTIDNNNNNTKLSYKCSVCNKEFSSYQALGGHKASHR 350
Query: 115 KLA-GAAAEDH-STSSAVTTSSASNGGGKV--HQCSICQKSFPTGQALGGHKRCHYEG-- 168
K + G +DH STSSA TTSSA+ GG V H+CSIC +SFPTGQALGGHKRCHYEG
Sbjct: 351 KNSVGGGGDDHPSTSSAATTSSANTNGGGVRSHECSICHRSFPTGQALGGHKRCHYEGVV 530
Query: 169 GGGASSTATATASEGVGSTHSHQRNFDLNLPAFPDFSASKFFVEEEVSSPLP---SKKPR 225
GGGAS+ T SEG+GSTHSHQR+FDLN+PAFP+F+ E+EV SP P KK R
Sbjct: 531 GGGASA---VTVSEGMGSTHSHQRDFDLNIPAFPEFANK--VGEDEVESPHPVMMKKKAR 695
Query: 226 --LLPKIEIPHY 235
++PKIEIP++
Sbjct: 696 VFVVPKIEIPNF 731
>TC86494 homologue to GP|7228329|emb|CAB77055.1 putative TFIIIA (or
kruppel)-like zinc finger protein {Medicago sativa},
partial (39%)
Length = 1092
Score = 158 bits (399), Expect = 2e-39
Identities = 105/256 (41%), Positives = 138/256 (53%), Gaps = 30/256 (11%)
Frame = +2
Query: 1 MAMEALKSPTTAAPTFTPFQEPNHSYMVDAP---WAKRKRSKRSRTDSHHNHASCTEEEY 57
+ ++ L S T A T P P + K+KRSKR R + TEEEY
Sbjct: 68 LELQTLNSSPTGATTTIPIPFPRFKQEENQERESLVKKKRSKRPRIGIGNPP---TEEEY 238
Query: 58 LALCLIMLARGSTAVTPKLTLSRPAPVTAEKLSYKCSVCEKTFPSYQALGGHKASHRKLA 117
LALCLIML++ + + + +P+ KL++KCSVC K FPSYQALGGHKASHRK
Sbjct: 239 LALCLIMLSQSNNQI-------QSSPL---KLNHKCSVCNKAFPSYQALGGHKASHRK-- 382
Query: 118 GAAAEDHSTSSAVTTSSASNGGGKVHQCSICQKSFPTGQALGGHKRCHYEG-------GG 170
++ ++ +++ T S S K+H+CSIC KSFPTGQALGGHKRCHYEG
Sbjct: 383 --SSSENQSTTVNETISVSVSTSKMHECSICHKSFPTGQALGGHKRCHYEGVINNNHNHN 556
Query: 171 GASSTATATASEGVGSTHSHQRNFDLNLPA----------FPDFSASKFFV--------E 212
++S+ + G S+ R FDLNLPA F + K V E
Sbjct: 557 NSNSSGITVSDAGAASSSISHRGFDLNLPAPLTEFWSPVGFGGGDSKKNKVNVNLAGINE 736
Query: 213 EEVSSPLP--SKKPRL 226
+EV SPLP +K+PRL
Sbjct: 737 QEVESPLPVTAKRPRL 784
>AW775690 similar to GP|8567776|gb|A hypothetical protein {Arabidopsis
thaliana}, partial (20%)
Length = 631
Score = 77.4 bits (189), Expect = 4e-15
Identities = 55/145 (37%), Positives = 64/145 (43%), Gaps = 33/145 (22%)
Frame = +2
Query: 54 EEEYLALCLIMLARGSTAVTP--KLTLSRPAPVTAEKLS----YKCSVCEKTFPSYQALG 107
EEE LA CLI+LARG +S K S Y+C C + FPS+QALG
Sbjct: 167 EEEDLANCLILLARGHNHRHDYQNTHVSNHRDHNNNKSSNFYLYECKTCNRCFPSFQALG 346
Query: 108 GHKASHRKLAGAAAEDHSTSSAVTTSS---------------------------ASNGGG 140
GH+ASH K A AVTTSS +
Sbjct: 347 GHRASHTKPNKANCNVAQQKQAVTTSSFVDDHYDPTMNTILSLQAFAATPITTIPTTKKS 526
Query: 141 KVHQCSICQKSFPTGQALGGHKRCH 165
KVH+CSIC F +GQALGGH R H
Sbjct: 527 KVHECSICGAEFSSGQALGGHMRRH 601
Score = 39.7 bits (91), Expect = 0.001
Identities = 22/71 (30%), Positives = 35/71 (48%)
Frame = +2
Query: 50 ASCTEEEYLALCLIMLARGSTAVTPKLTLSRPAPVTAEKLSYKCSVCEKTFPSYQALGGH 109
+S ++ Y +L+ + A TP T+ P T + ++CS+C F S QALGGH
Sbjct: 422 SSFVDDHYDPTMNTILSLQAFAATPITTI----PTTKKSKVHECSICGAEFSSGQALGGH 589
Query: 110 KASHRKLAGAA 120
H+ L +
Sbjct: 590 MRRHKNLVNTS 622
>BG648193 similar to GP|2346976|dbj| ZPT2-13 {Petunia x hybrida}, partial
(43%)
Length = 629
Score = 77.4 bits (189), Expect = 4e-15
Identities = 45/110 (40%), Positives = 62/110 (55%), Gaps = 2/110 (1%)
Frame = +2
Query: 58 LALCLIMLARGSTAVTPKLTLSRPAPVTAEKLSYKCSVCEKTFPSYQALGGHKASHR--K 115
LA CL+ML+ ++P + + Y+C C K FPS+QALGGH+ASH+ K
Sbjct: 134 LANCLMMLSYPQHQPQN----NKPNQKSFAPVEYECKTCNKKFPSFQALGGHRASHKRSK 301
Query: 116 LAGAAAEDHSTSSAVTTSSASNGGGKVHQCSICQKSFPTGQALGGHKRCH 165
L G +STS ++ K+H+CSIC ++F GQALGGH R H
Sbjct: 302 LEGDELLTNSTSLSLGNKP------KMHECSICGQNFSLGQALGGHMRRH 433
Score = 38.5 bits (88), Expect = 0.002
Identities = 25/81 (30%), Positives = 35/81 (42%), Gaps = 6/81 (7%)
Frame = +2
Query: 143 HQCSICQKSFPTGQALGGHKRCHYEGGGGASSTATATASEGVGS-THSHQ-----RNFDL 196
++C C K FP+ QALGGH+ H T + S +G+ H+ +NF L
Sbjct: 221 YECKTCNKKFPSFQALGGHRASHKRSKLEGDELLTNSTSLSLGNKPKMHECSICGQNFSL 400
Query: 197 NLPAFPDFSASKFFVEEEVSS 217
K + EEVSS
Sbjct: 401 GQALGGHMRRHKAIMNEEVSS 463
Score = 30.8 bits (68), Expect = 0.47
Identities = 11/26 (42%), Positives = 17/26 (65%)
Frame = +2
Query: 91 YKCSVCEKTFPSYQALGGHKASHRKL 116
++CS+C + F QALGGH H+ +
Sbjct: 365 HECSICGQNFSLGQALGGHMRRHKAI 442
>TC88075 homologue to GP|2346978|dbj|BAA21923.1 ZPT2-14 {Petunia x hybrida},
partial (38%)
Length = 674
Score = 74.7 bits (182), Expect = 3e-14
Identities = 38/92 (41%), Positives = 52/92 (56%)
Frame = +3
Query: 85 TAEKLSYKCSVCEKTFPSYQALGGHKASHRKLAGAAAEDHSTSSAVTTSSASNGGGKVHQ 144
T + ++C C + F S+QALGGH+ASH+KL +E+ + N K+HQ
Sbjct: 126 TYSPVEFECKTCNRKFSSFQALGGHRASHKKLKLLDSEE------AHKVNIHNNKPKMHQ 287
Query: 145 CSICQKSFPTGQALGGHKRCHYEGGGGASSTA 176
CSIC + F GQALGGH R H G SS++
Sbjct: 288 CSICGQEFKLGQALGGHMRRHRINNEGFSSSS 383
>TC81612 similar to GP|2346978|dbj|BAA21923.1 ZPT2-14 {Petunia x hybrida},
partial (50%)
Length = 612
Score = 73.6 bits (179), Expect = 6e-14
Identities = 38/90 (42%), Positives = 51/90 (56%)
Frame = +3
Query: 76 LTLSRPAPVTAEKLSYKCSVCEKTFPSYQALGGHKASHRKLAGAAAEDHSTSSAVTTSSA 135
+ LS P + E Y+C C K F S+QALGGH+ASH+++ A E+ + S +
Sbjct: 147 MLLSCPQQKSYENGEYECKTCNKKFSSFQALGGHRASHKRMKLAEGEELKEQA---KSLS 317
Query: 136 SNGGGKVHQCSICQKSFPTGQALGGHKRCH 165
K+H+CSIC F GQALGGH R H
Sbjct: 318 LWNKPKMHECSICGMGFSLGQALGGHMRKH 407
Score = 30.4 bits (67), Expect = 0.62
Identities = 12/24 (50%), Positives = 15/24 (62%)
Frame = +3
Query: 91 YKCSVCEKTFPSYQALGGHKASHR 114
++CS+C F QALGGH HR
Sbjct: 339 HECSICGMGFSLGQALGGHMRKHR 410
>TC88653 similar to GP|1786134|dbj|BAA19110.1 PEThy;ZPT2-5 {Petunia x
hybrida}, partial (59%)
Length = 866
Score = 72.4 bits (176), Expect = 1e-13
Identities = 52/155 (33%), Positives = 75/155 (47%), Gaps = 4/155 (2%)
Frame = +1
Query: 39 KRSRTDSHHNHASCTEEEYLALCLIMLARGSTAVTPKLTLSRPAPVTAEKLSYKCSVCEK 98
KR R ++ H S T +YL L L+ GS + ++ S ++C C++
Sbjct: 22 KRER-ETDHTTNSITMAKYLML----LSGGSDKIFDQVNYSS----NFNNRVFECKTCKR 174
Query: 99 TFPSYQALGGHKASHRK----LAGAAAEDHSTSSAVTTSSASNGGGKVHQCSICQKSFPT 154
F S+QALGGH+ASH+K + +DH S +T+ A K H CSIC F
Sbjct: 175 QFSSFQALGGHRASHKKPRLMEMTSDGDDHHGSILTSTTKA-----KTHACSICGLEFGI 339
Query: 155 GQALGGHKRCHYEGGGGASSTATATASEGVGSTHS 189
GQALGGH R H T ++ A+ G+ H+
Sbjct: 340 GQALGGHMRRH-------RRTESSKANNSNGNMHN 423
>TC81921 homologue to GP|2346978|dbj|BAA21923.1 ZPT2-14 {Petunia x hybrida},
partial (41%)
Length = 839
Score = 72.0 bits (175), Expect = 2e-13
Identities = 47/139 (33%), Positives = 65/139 (45%), Gaps = 7/139 (5%)
Frame = +3
Query: 58 LALCLIMLARGSTAVTPKLTLSRPAPVTAEKLSYKCSVCEKTFPSYQALGGHKASHRK-- 115
+A CL++L+RGS + + T+ ++C C + FPS+QALGGH+ASH+K
Sbjct: 159 MANCLMLLSRGSDQFEATYSST-----TSNNRVFECKTCNRQFPSFQALGGHRASHKKPR 323
Query: 116 LAGAAAEDHSTSSAVTTSSASNGGGKVHQCSICQKSFPTGQALGGHKRCHYEGGGGA--- 172
L G + + K H+CSIC F GQALGGH R H
Sbjct: 324 LMGENIDGQLLHTPPKP--------KTHECSICGLEFAIGQALGGHMRRHRAANMNGNKN 479
Query: 173 --SSTATATASEGVGSTHS 189
+S T + S G G S
Sbjct: 480 MHNSNNTMSCSSGGGGDSS 536
>TC93387 similar to GP|1786136|dbj|BAA19111.1 PEThy;ZPT2-6 {Petunia x
hybrida}, partial (17%)
Length = 650
Score = 53.9 bits (128), Expect = 5e-08
Identities = 32/76 (42%), Positives = 41/76 (53%), Gaps = 14/76 (18%)
Frame = +1
Query: 54 EEEYLALCLIMLARGSTAVTPKLTLSRPAPV-----------TAEKLS---YKCSVCEKT 99
EEE LA CLI+LA+G + V T+ KL Y+C C +T
Sbjct: 319 EEEDLAKCLILLAQGGNHREDGGVVDENKRVKGSHGNKKIGETSTKLGLYIYECKTCNRT 498
Query: 100 FPSYQALGGHKASHRK 115
FPS+QALGGH+ASH+K
Sbjct: 499 FPSFQALGGHRASHKK 546
>AL371585 similar to PIR|S69193|S691 probable finger protein Pszf1 - garden
pea, partial (19%)
Length = 497
Score = 50.4 bits (119), Expect = 6e-07
Identities = 38/96 (39%), Positives = 48/96 (49%), Gaps = 2/96 (2%)
Frame = +1
Query: 134 SASNGG-GKVHQCSICQKSFPTGQALGGHKRCHYEGGGGASSTATATASEGVGSTHSHQR 192
+A NG K+H+CSIC F +GQALGGH R H A++ A A E V + Q
Sbjct: 40 AAINGNKAKIHECSICGSEFTSGQALGGHMRRHRV--SVANAAAVAAPDERVRPRNILQ- 210
Query: 193 NFDLNLPA-FPDFSASKFFVEEEVSSPLPSKKPRLL 227
DLNLPA D SKF S + + P L+
Sbjct: 211 -LDLNLPAPEEDIRESKFQFPATQKSMVMAAAPALV 315
Score = 35.0 bits (79), Expect = 0.025
Identities = 17/32 (53%), Positives = 21/32 (65%), Gaps = 1/32 (3%)
Frame = +1
Query: 91 YKCSVCEKTFPSYQALGGHKASHR-KLAGAAA 121
++CS+C F S QALGGH HR +A AAA
Sbjct: 70 HECSICGSEFTSGQALGGHMRRHRVSVANAAA 165
>TC82775 similar to PIR|T52382|T52382 zinc finger protein ZPT4-4 C2H2-type
[imported] - garden petunia, partial (17%)
Length = 1152
Score = 45.4 bits (106), Expect = 2e-05
Identities = 28/113 (24%), Positives = 45/113 (39%), Gaps = 32/113 (28%)
Frame = +3
Query: 87 EKLSYKCSVCEKTFPSYQALGGHKASHRKLAGAAAEDHST---------SSAVTTSSASN 137
++ + C C K+FP ++LGGH SH A +H + ++ + +A+N
Sbjct: 138 QEFKHACKFCSKSFPCGRSLGGHMRSHINNLSAEKAEHKEKQLIFSTKKNDSIGSEAATN 317
Query: 138 GGGKVHQ-----------------------CSICQKSFPTGQALGGHKRCHYE 167
G + + C C K F + QAL GH +CH E
Sbjct: 318 GSYGLRENPKKTWRKRIADSSEDSYVYDKFCKECGKCFHSWQALFGHMKCHSE 476
>BI308410 similar to PIR|S55882|S558 CCHH finger protein 2 - Arabidopsis
thaliana, partial (46%)
Length = 492
Score = 40.8 bits (94), Expect = 5e-04
Identities = 24/81 (29%), Positives = 38/81 (46%)
Frame = +3
Query: 87 EKLSYKCSVCEKTFPSYQALGGHKASHRKLAGAAAEDHSTSSAVTTSSASNGGGKVHQCS 146
E+ + C+ C++ F S QALGGH+ +H+ A + SSA+ +S A +
Sbjct: 90 EQRIFSCNYCQRKFYSSQALGGHQNAHKLERTLAKKSREMSSAMQSSYA--------ELP 245
Query: 147 ICQKSFPTGQALGGHKRCHYE 167
+F T LG H H +
Sbjct: 246 EHPSNFSTNYHLGSHGNAHLD 308
Score = 38.1 bits (87), Expect = 0.003
Identities = 15/38 (39%), Positives = 26/38 (67%)
Frame = +3
Query: 128 SAVTTSSASNGGGKVHQCSICQKSFPTGQALGGHKRCH 165
S+ ++SS+S+ ++ C+ CQ+ F + QALGGH+ H
Sbjct: 57 SSSSSSSSSSMEQRIFSCNYCQRKFYSSQALGGHQNAH 170
>AW686309 homologue to PIR|S55886|S558 CCHH finger protein 6 - Arabidopsis
thaliana, partial (25%)
Length = 669
Score = 37.7 bits (86), Expect = 0.004
Identities = 18/53 (33%), Positives = 28/53 (51%), Gaps = 2/53 (3%)
Frame = +2
Query: 115 KLAGAAAEDHSTSSAVTTSSASN--GGGKVHQCSICQKSFPTGQALGGHKRCH 165
KL G ++ + + T S +S G K+++C C + F QALGGH+ H
Sbjct: 137 KLFGINIQEACLNQSPTQSESSEPLNGRKLYECQYCCREFANSQALGGHQNAH 295
Score = 35.0 bits (79), Expect = 0.025
Identities = 15/33 (45%), Positives = 22/33 (66%)
Frame = +2
Query: 83 PVTAEKLSYKCSVCEKTFPSYQALGGHKASHRK 115
P+ KL Y+C C + F + QALGGH+ +H+K
Sbjct: 206 PLNGRKL-YECQYCCREFANSQALGGHQNAHKK 301
>BI310179 similar to PIR|T52381|T523 zinc finger protein Zat1 C2H2-type
[imported] - Arabidopsis thaliana, partial (16%)
Length = 704
Score = 37.0 bits (84), Expect = 0.007
Identities = 13/23 (56%), Positives = 16/23 (69%)
Frame = +2
Query: 143 HQCSICQKSFPTGQALGGHKRCH 165
H+C C +SF G+ALGGH R H
Sbjct: 77 HKCKFCMRSFSNGRALGGHMRSH 145
Score = 30.4 bits (67), Expect = 0.62
Identities = 11/23 (47%), Positives = 16/23 (68%)
Frame = +2
Query: 91 YKCSVCEKTFPSYQALGGHKASH 113
+KC C ++F + +ALGGH SH
Sbjct: 77 HKCKFCMRSFSNGRALGGHMRSH 145
>BG449113 similar to PIR|S55881|S558 CCHH finger protein 1 - Arabidopsis
thaliana, partial (15%)
Length = 632
Score = 36.6 bits (83), Expect = 0.009
Identities = 20/55 (36%), Positives = 30/55 (54%), Gaps = 2/55 (3%)
Frame = +1
Query: 85 TAEKLSYKCSVCEKTFPSYQALGGHKASHRKLAGAAAEDHST--SSAVTTSSASN 137
T+E + C+ C++ F S QALGGH+ +H++ A T S+ TTS N
Sbjct: 211 TSEPRFFTCNYCKRKFFSSQALGGHQNAHKRERSIAKRGRRTMFSATGTTSFLHN 375
Score = 35.0 bits (79), Expect = 0.025
Identities = 20/72 (27%), Positives = 31/72 (42%), Gaps = 4/72 (5%)
Frame = +1
Query: 125 STSSAVTTSSASNGGGKVHQCSICQKSFPTGQALGGHKRCHYE----GGGGASSTATATA 180
++S T S S + C+ C++ F + QALGGH+ H G + +AT
Sbjct: 175 TSSENFTCGSESTSEPRFFTCNYCKRKFFSSQALGGHQNAHKRERSIAKRGRRTMFSATG 354
Query: 181 SEGVGSTHSHQR 192
+ H H R
Sbjct: 355 TTSFLHNHLHHR 390
>TC82590 similar to PIR|T50806|T50806 zinc finger-like protein - Arabidopsis
thaliana, partial (19%)
Length = 690
Score = 36.6 bits (83), Expect = 0.009
Identities = 26/101 (25%), Positives = 41/101 (39%), Gaps = 13/101 (12%)
Frame = +2
Query: 87 EKLSYKCSVCEKTFPSYQALGGHKASHRKLAGAAAEDHSTSSAVTTSSASNGGGKVH--Q 144
E+ + C+ C + F S QALGGH+ +H++ A + + S+++ G +H Q
Sbjct: 323 EQRVFSCNYCHRKFYSSQALGGHQNAHKRERSIAKRGQRFGTQIIASASAFGFPILHNKQ 502
Query: 145 CSICQKSFPTGQALGG-----------HKRCHYEGGGGASS 174
S P A G HK H G +S
Sbjct: 503 SYANMASLPLYGAFGNKPLGIQAHSMIHKPSHVSSNGFGNS 625
>TC82737 homologue to PIR|B96686|B96686 probable C2H2-type zinc finger
protein F15E12.19 [imported] - Arabidopsis thaliana,
partial (18%)
Length = 1006
Score = 36.2 bits (82), Expect = 0.011
Identities = 16/52 (30%), Positives = 31/52 (58%)
Frame = +2
Query: 64 MLARGSTAVTPKLTLSRPAPVTAEKLSYKCSVCEKTFPSYQALGGHKASHRK 115
++ R ST ++ T +T +++ + C+ C + F S QALGGH+ +H++
Sbjct: 335 LVVRDSTGISFSSTSESSNELTIQRV-FSCNYCRRKFYSSQALGGHQNAHKR 487
Score = 33.5 bits (75), Expect = 0.073
Identities = 15/44 (34%), Positives = 24/44 (54%), Gaps = 5/44 (11%)
Frame = +2
Query: 127 SSAVTTSSASNGGG-----KVHQCSICQKSFPTGQALGGHKRCH 165
S+ ++ SS S +V C+ C++ F + QALGGH+ H
Sbjct: 350 STGISFSSTSESSNELTIQRVFSCNYCRRKFYSSQALGGHQNAH 481
>TC88893 similar to GP|6478938|gb|AAF14043.1| putative C2H2-type zinc finger
protein {Arabidopsis thaliana}, partial (25%)
Length = 819
Score = 35.8 bits (81), Expect = 0.015
Identities = 18/54 (33%), Positives = 30/54 (55%), Gaps = 4/54 (7%)
Frame = +1
Query: 90 SYKCSVCEKTFPSYQALGGHKASHR----KLAGAAAEDHSTSSAVTTSSASNGG 139
SY C+ C++ F + QALGGH HR KL ++E++ S + + ++ G
Sbjct: 127 SYSCTFCKRGFSNAQALGGHMNIHRRDRAKLKQQSSEENLLSLDIANKNPNDQG 288
Score = 34.3 bits (77), Expect = 0.043
Identities = 14/44 (31%), Positives = 23/44 (51%)
Frame = +1
Query: 122 EDHSTSSAVTTSSASNGGGKVHQCSICQKSFPTGQALGGHKRCH 165
++ + S + SS G + + C+ C++ F QALGGH H
Sbjct: 67 QNPNKSDQIRWSSDDPGQVRSYSCTFCKRGFSNAQALGGHMNIH 198
>TC88476 homologue to GP|21592423|gb|AAM64374.1 zinc finger protein 5 ZFP5
{Arabidopsis thaliana}, partial (21%)
Length = 945
Score = 35.8 bits (81), Expect = 0.015
Identities = 22/63 (34%), Positives = 28/63 (43%), Gaps = 15/63 (23%)
Frame = +2
Query: 118 GAAAEDHSTSSAVTTS-SASNGGGKVHQ--------------CSICQKSFPTGQALGGHK 162
G A E + +V +S S S+GG K+ Q C C K F QALGGH+
Sbjct: 128 GVAKESNEGDESVNSSNSISSGGDKIVQEKNSSKDQDERKFECQYCFKEFANSQALGGHQ 307
Query: 163 RCH 165
H
Sbjct: 308 NAH 316
Score = 33.9 bits (76), Expect = 0.056
Identities = 13/41 (31%), Positives = 24/41 (57%)
Frame = +2
Query: 75 KLTLSRPAPVTAEKLSYKCSVCEKTFPSYQALGGHKASHRK 115
K+ + + ++ ++C C K F + QALGGH+ +H+K
Sbjct: 200 KIVQEKNSSKDQDERKFECQYCFKEFANSQALGGHQNAHKK 322
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.313 0.126 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,441,835
Number of Sequences: 36976
Number of extensions: 130228
Number of successful extensions: 766
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 681
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 722
length of query: 236
length of database: 9,014,727
effective HSP length: 93
effective length of query: 143
effective length of database: 5,575,959
effective search space: 797362137
effective search space used: 797362137
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0050.4


