

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0050.12
(143 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
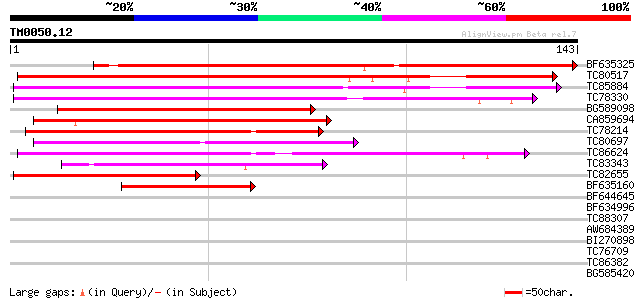
Score E
Sequences producing significant alignments: (bits) Value
BF635325 similar to GP|4204575|gb|A cytochrome b5 DIF-F {Petunia... 161 7e-41
TC80517 similar to PIR|S46306|S46306 cytochrome b5 - common toba... 109 3e-25
TC85884 similar to GP|2695711|emb|CAA04703.1 cytochome b5 {Olea ... 97 3e-21
TC78330 similar to GP|2695711|emb|CAA04703.1 cytochome b5 {Olea ... 90 3e-19
BG589098 similar to GP|21536989|gb| cytochrome b5 {Arabidopsis t... 80 3e-16
CA859694 weakly similar to GP|13325120|gb| Unknown (protein for ... 72 8e-14
TC78214 similar to SP|P39869|NIA_LOTJA Nitrate reductase (EC 1.6... 72 1e-13
TC80697 similar to PIR|H96631|H96631 probable Cytochrome B5 F8A5... 59 7e-10
TC86624 similar to PIR|A84900|A84900 hypothetical protein At2g46... 52 6e-08
TC83343 weakly similar to GP|1783355|emb|CAA71448.1 delta-9 fatt... 44 2e-05
TC82655 similar to PIR|T49930|T49930 hypothetical protein F17I14... 42 6e-05
BF635160 similar to SP|Q42342|CY51_ Cytochrome b5 isoform 1. [Mo... 42 8e-05
BF644645 30 0.25
BF634996 similar to GP|2695711|emb| cytochome b5 {Olea europaea}... 30 0.43
TC88307 homologue to GP|6782273|emb|CAB70255.1 contains similari... 28 1.2
AW684389 similar to GP|9759091|dbj contains similarity to unknow... 28 1.2
BI270898 28 1.6
TC76709 similar to GP|20259017|gb|AAM14224.1 unknown protein {Ar... 27 2.8
TC86382 similar to PIR|C84642|C84642 probable steroid binding pr... 27 3.6
BG585420 similar to GP|17104707|gb unknown protein {Arabidopsis ... 26 4.7
>BF635325 similar to GP|4204575|gb|A cytochrome b5 DIF-F {Petunia x hybrida},
partial (35%)
Length = 682
Score = 161 bits (408), Expect = 7e-41
Identities = 83/126 (65%), Positives = 101/126 (79%), Gaps = 4/126 (3%)
Frame = +1
Query: 22 WVVINGRVLDVTKFLIEHPGGDDVLLEVAGKDVTKEFDAVGHSKEAQNLVLKYQVGILQG 81
WV++ +VLDVTKFL EHPGG++V++EVAGKD TKEFDAVGHSK AQNLVLKYQVG+L+G
Sbjct: 145 WVLV--QVLDVTKFLEEHPGGEEVIVEVAGKDATKEFDAVGHSKVAQNLVLKYQVGVLEG 318
Query: 82 ATVQEID----VVHKEPNSKQEMSAFVIKEDAESKSVALFEFFVPLVVATLYFGYRCLTR 137
ATV+++D V EP SK EMSAFVIKED+ SK+V EFFVP++ A +YFGYR +T
Sbjct: 319 ATVEKVDGKDVVEDNEPRSK-EMSAFVIKEDSTSKTVTFLEFFVPIIFACIYFGYRLITI 495
Query: 138 TDPVTY 143
D V Y
Sbjct: 496 ADTVDY 513
>TC80517 similar to PIR|S46306|S46306 cytochrome b5 - common tobacco,
partial (94%)
Length = 741
Score = 109 bits (273), Expect = 3e-25
Identities = 58/139 (41%), Positives = 88/139 (62%), Gaps = 3/139 (2%)
Frame = +3
Query: 3 ERRVFTLSQVAQHKSNKDCWVVINGRVLDVTKFLIEHPGGDDVLLEVAGKDVTKEFDAVG 62
ER+VFTL+ V++H S KDCW+VI+ +V DVTKFL +HPGGD+VL+ GKD + +FD +G
Sbjct: 84 ERKVFTLADVSKHNSAKDCWLVIHNKVYDVTKFLEDHPGGDEVLISSTGKDASNDFDDIG 263
Query: 63 HSKEAQNLVLKYQVGILQGATV-QEIDVV-HKEPNSKQE-MSAFVIKEDAESKSVALFEF 119
HS A ++ +Y VG + +T+ ++D K+P+ Q+ S FVIK + +F
Sbjct: 264 HSTSAYTMMEEYYVGDIDSSTIPSKVDYTPPKQPHYNQDKTSEFVIK---------ILQF 416
Query: 120 FVPLVVATLYFGYRCLTRT 138
VPL + + G R T++
Sbjct: 417 LVPLFILGVAVGIRFYTKS 473
>TC85884 similar to GP|2695711|emb|CAA04703.1 cytochome b5 {Olea europaea},
partial (98%)
Length = 708
Score = 96.7 bits (239), Expect = 3e-21
Identities = 50/142 (35%), Positives = 82/142 (57%), Gaps = 4/142 (2%)
Frame = +1
Query: 2 AERRVFTLSQVAQHKSNKDCWVVINGRVLDVTKFLIEHPGGDDVLLEVAGKDVTKEFDAV 61
++ ++ T V++H KDCW++++G+V DV+ F+ +HPGGD+VLL GKD T +F+ V
Sbjct: 88 SDPKLHTFDDVSKHNKTKDCWLILSGKVYDVSPFMEDHPGGDEVLLSATGKDATNDFEDV 267
Query: 62 GHSKEAQNLVLKYQVGILQGATVQEIDVVHKEPNSKQ----EMSAFVIKEDAESKSVALF 117
GHS A+ ++ KY +G + +TV + + P Q + S FVIK +
Sbjct: 268 GHSDSAREMMDKYYIGEIDPSTV-PLKRTYVPPQQSQYNPDKTSEFVIK---------IL 417
Query: 118 EFFVPLVVATLYFGYRCLTRTD 139
+F VPL++ L F R T+ +
Sbjct: 418 QFLVPLLILGLAFVVRNYTKKE 483
>TC78330 similar to GP|2695711|emb|CAA04703.1 cytochome b5 {Olea europaea},
partial (70%)
Length = 724
Score = 89.7 bits (221), Expect = 3e-19
Identities = 49/138 (35%), Positives = 71/138 (50%), Gaps = 6/138 (4%)
Frame = +3
Query: 2 AERRVFTLSQVAQHKSNKDCWVVINGRVLDVTKFLIEHPGGDDVLLEVAGKDVTKEFDAV 61
++ T VA+H DCW+++N +V DVT FL +HPGGD+ LL GKD T +F+ V
Sbjct: 57 SKSETLTFEDVAKHNHKNDCWIIVNKKVYDVTPFLDDHPGGDEALLSATGKDATTDFEDV 236
Query: 62 GHSKEAQNLVLKYQVGILQGATVQEIDVVHKEPNSKQEMSAFVIKEDAESKSVALF-EFF 120
GHS A ++ KY VG T+ V N + A I + S + F ++
Sbjct: 237 GHSDSATEMMEKYYVGEFDANTLP----VEARNNPTAPIQASTINSNQSSGVLLKFLQYL 404
Query: 121 VPLVV-----ATLYFGYR 133
VPL++ A Y+G R
Sbjct: 405 VPLLILGVAFALQYYGKR 458
>BG589098 similar to GP|21536989|gb| cytochrome b5 {Arabidopsis thaliana},
partial (48%)
Length = 280
Score = 80.1 bits (196), Expect = 3e-16
Identities = 30/65 (46%), Positives = 49/65 (75%)
Frame = +2
Query: 13 AQHKSNKDCWVVINGRVLDVTKFLIEHPGGDDVLLEVAGKDVTKEFDAVGHSKEAQNLVL 72
+ HK+ DCW++++G+V DVT++ + PGGDDV+L+ G+D T++F+ GHSK A+ L+
Sbjct: 2 SNHKTKDDCWIIVDGKVYDVTQYSDDPPGGDDVILDATGRDATEDFEDAGHSKSARELME 181
Query: 73 KYQVG 77
KY +G
Sbjct: 182KYYIG 196
>CA859694 weakly similar to GP|13325120|gb| Unknown (protein for MGC:10477)
{Homo sapiens}, partial (41%)
Length = 277
Score = 72.0 bits (175), Expect = 8e-14
Identities = 33/78 (42%), Positives = 53/78 (67%), Gaps = 3/78 (3%)
Frame = +1
Query: 7 FTLSQVAQH---KSNKDCWVVINGRVLDVTKFLIEHPGGDDVLLEVAGKDVTKEFDAVGH 63
FT S++++ K NK + V+NG+V+DVTKF EHPGG++VL+ G D T F+++GH
Sbjct: 28 FTYSELSEKYNGKDNKPLYFVVNGKVVDVTKFKDEHPGGEEVLMGEGGLDATDSFESIGH 207
Query: 64 SKEAQNLVLKYQVGILQG 81
S EA+ ++ ++G + G
Sbjct: 208 SDEAREILKSLEIGDVVG 261
>TC78214 similar to SP|P39869|NIA_LOTJA Nitrate reductase (EC 1.6.6.1) (NR).
{Lotus japonicus}, partial (65%)
Length = 2021
Score = 71.6 bits (174), Expect = 1e-13
Identities = 32/75 (42%), Positives = 52/75 (68%)
Frame = +2
Query: 5 RVFTLSQVAQHKSNKDCWVVINGRVLDVTKFLIEHPGGDDVLLEVAGKDVTKEFDAVGHS 64
+ ++LS+V +HK+ W++++G V D T+FL +HPGG D +L AG D T EF+A+ HS
Sbjct: 683 KTYSLSEVKKHKTLDSAWIIVHGNVYDCTRFLKDHPGGSDSILINAGTDCTDEFEAI-HS 859
Query: 65 KEAQNLVLKYQVGIL 79
+A+ L+ Y++G L
Sbjct: 860 DKAKKLLEDYRIGEL 904
>TC80697 similar to PIR|H96631|H96631 probable Cytochrome B5 F8A5.18
[imported] - Arabidopsis thaliana, partial (61%)
Length = 905
Score = 58.9 bits (141), Expect = 7e-10
Identities = 26/82 (31%), Positives = 47/82 (56%)
Frame = +2
Query: 7 FTLSQVAQHKSNKDCWVVINGRVLDVTKFLIEHPGGDDVLLEVAGKDVTKEFDAVGHSKE 66
++ ++V H + DCW++I +V DVT ++ EHPGGD +L AG D T+ F H+
Sbjct: 242 YSKTEVTLHNTRTDCWIIIKNKVYDVTSYVEEHPGGDAILAH-AGDDSTEGFFGPQHATR 418
Query: 67 AQNLVLKYQVGILQGATVQEID 88
+++ + +G L+ + +D
Sbjct: 419 VFDMIEDFYIGDLEQ*PILHLD 484
>TC86624 similar to PIR|A84900|A84900 hypothetical protein At2g46210
[imported] - Arabidopsis thaliana, partial (98%)
Length = 2039
Score = 52.4 bits (124), Expect = 6e-08
Identities = 39/131 (29%), Positives = 67/131 (50%), Gaps = 2/131 (1%)
Frame = +3
Query: 3 ERRVFTLSQVAQHKSNKDCWVVINGRVLDVTKFLIEHPGGDDVLLEVAGKDVTKEFDAVG 62
+++ T ++ +H D W+ I G+V +V+ + +HPGG+ VLL +AG+DVT F A
Sbjct: 483 KKKYITSEELKKHDKEGDLWISIQGKVYNVSDWAKKHPGGEVVLLNLAGQDVTDAFIAY- 659
Query: 63 HSKEAQNLVLKYQVGILQGATVQEIDVVHKEPNSKQEMSAFVIKEDAESKS-VALFEF-F 120
H A K+ G +++ V + ++ +S FV E K VALF
Sbjct: 660 HPGSA----WKHLDQFFTGYYLEDFKVSEVSKDYRRLVSEFVKMGLFEKKEHVALFTLTS 827
Query: 121 VPLVVATLYFG 131
V +++A + +G
Sbjct: 828 VAIMLAIVVYG 860
>TC83343 weakly similar to GP|1783355|emb|CAA71448.1 delta-9 fatty acid
desaturase {Cryptococcus curvatus}, partial (14%)
Length = 682
Score = 44.3 bits (103), Expect = 2e-05
Identities = 24/69 (34%), Positives = 38/69 (54%), Gaps = 2/69 (2%)
Frame = +1
Query: 14 QHKSNKDCWVVINGRVLDVTKFLIEHPGGDDVLLEVAGKDVTKEF--DAVGHSKEAQNLV 71
Q + K+ W++++ VLDV+ F +HPGG +L GKD TK F HSK A +
Sbjct: 337 QEQEGKE-WILVDEFVLDVSSFKDDHPGGAKILKNYYGKDSTKAFYGGLNDHSKAANTMT 513
Query: 72 LKYQVGILQ 80
++V ++
Sbjct: 514 AMFRVAKIE 540
>TC82655 similar to PIR|T49930|T49930 hypothetical protein F17I14.130 -
Arabidopsis thaliana, partial (45%)
Length = 719
Score = 42.4 bits (98), Expect = 6e-05
Identities = 15/47 (31%), Positives = 32/47 (67%)
Frame = +1
Query: 2 AERRVFTLSQVAQHKSNKDCWVVINGRVLDVTKFLIEHPGGDDVLLE 48
+ RR+ ++ +V +HK+ + W V+ GRV +++ ++ HP G D+L++
Sbjct: 571 SNRRLISMDEVRKHKTEGEMWTVLKGRVYNISPYMKFHPEGVDMLMK 711
>BF635160 similar to SP|Q42342|CY51_ Cytochrome b5 isoform 1. [Mouse-ear
cress] {Arabidopsis thaliana}, partial (38%)
Length = 688
Score = 42.0 bits (97), Expect = 8e-05
Identities = 18/34 (52%), Positives = 23/34 (66%)
Frame = +1
Query: 29 VLDVTKFLIEHPGGDDVLLEVAGKDVTKEFDAVG 62
V DV+ F+ +HPGGD LL GKD T +F+ VG
Sbjct: 586 VYDVSPFMEDHPGGDXGLLSATGKDATNDFEDVG 687
Score = 27.7 bits (60), Expect = 1.6
Identities = 7/17 (41%), Positives = 14/17 (82%)
Frame = +3
Query: 12 VAQHKSNKDCWVVINGR 28
V++H KDCW++++G+
Sbjct: 87 VSKHNKTKDCWLILSGK 137
>BF644645
Length = 676
Score = 30.4 bits (67), Expect = 0.25
Identities = 9/28 (32%), Positives = 16/28 (57%)
Frame = +2
Query: 2 AERRVFTLSQVAQHKSNKDCWVVINGRV 29
++ T VA+H DCW+++N +V
Sbjct: 38 SKSETLTFEDVAKHNHKNDCWIIVNKKV 121
>BF634996 similar to GP|2695711|emb| cytochome b5 {Olea europaea}, partial
(17%)
Length = 660
Score = 29.6 bits (65), Expect = 0.43
Identities = 9/23 (39%), Positives = 16/23 (69%)
Frame = +1
Query: 12 VAQHKSNKDCWVVINGRVLDVTK 34
V++H KDCW++++G+V K
Sbjct: 100 VSKHNKTKDCWLILSGKVFFFAK 168
>TC88307 homologue to GP|6782273|emb|CAB70255.1 contains similarity to Pfam
domain: PF01748 (Domain of unknown function)
Score=122.0 E-value=3., partial (4%)
Length = 1681
Score = 28.1 bits (61), Expect = 1.2
Identities = 15/57 (26%), Positives = 27/57 (47%)
Frame = +2
Query: 66 EAQNLVLKYQVGILQGATVQEIDVVHKEPNSKQEMSAFVIKEDAESKSVALFEFFVP 122
+ Q + Q+ I + AT+Q++DV + SK + +DA K+ E +P
Sbjct: 548 DVQIAIASPQIAITKPATIQDVDVKICQETSKANDDQVALNDDAVMKASTNIEQHLP 718
>AW684389 similar to GP|9759091|dbj contains similarity to unknown
protein~gb|AAF47871.1~gene_id:MJJ3.6 {Arabidopsis
thaliana}, partial (16%)
Length = 656
Score = 28.1 bits (61), Expect = 1.2
Identities = 16/47 (34%), Positives = 26/47 (55%)
Frame = +2
Query: 89 VVHKEPNSKQEMSAFVIKEDAESKSVALFEFFVPLVVATLYFGYRCL 135
V KEP Q+M + +K + LF FF+ ++VA ++GY+ L
Sbjct: 377 VESKEPTILQKMIS-------RAKQLLLFIFFLVILVAATHYGYKGL 496
>BI270898
Length = 653
Score = 27.7 bits (60), Expect = 1.6
Identities = 14/50 (28%), Positives = 27/50 (54%), Gaps = 6/50 (12%)
Frame = -1
Query: 98 QEMSAFVIKEDAESKSVALFEFFVPLVVAT------LYFGYRCLTRTDPV 141
Q A + ++A++ S +LF+FFVP+ + LY ++ + RT +
Sbjct: 278 QNSEALTLYKEAKTDSSSLFKFFVPITIKINSIHQYLYILHKNIKRTQQI 129
>TC76709 similar to GP|20259017|gb|AAM14224.1 unknown protein {Arabidopsis
thaliana}, partial (87%)
Length = 2358
Score = 26.9 bits (58), Expect = 2.8
Identities = 18/51 (35%), Positives = 28/51 (54%), Gaps = 3/51 (5%)
Frame = -3
Query: 95 NSKQEMSAFVIK---EDAESKSVALFEFFVPLVVATLYFGYRCLTRTDPVT 142
+S+ E SA ++ DA SKS LF+ PL+ T G+ C++R+ T
Sbjct: 1891 SSESETSARSLQALASDASSKSFFLFKTMCPLISHTA--GFTCVSRSSAST 1745
>TC86382 similar to PIR|C84642|C84642 probable steroid binding protein
[imported] - Arabidopsis thaliana, complete
Length = 797
Score = 26.6 bits (57), Expect = 3.6
Identities = 26/93 (27%), Positives = 44/93 (46%), Gaps = 4/93 (4%)
Frame = +3
Query: 3 ERRVFTLSQVAQHKS---NKDCWVVINGRVLDVTKFLIEH-PGGDDVLLEVAGKDVTKEF 58
E T Q++++ +K +V + GRV DVT + PGG + AGKD ++
Sbjct: 81 EEMEMTAQQLSEYNGTDPSKPIYVAVKGRVYDVTTGKSFYGPGGAYAMF--AGKDASRAL 254
Query: 59 DAVGHSKEAQNLVLKYQVGILQGATVQEIDVVH 91
+ SK +++ L G T +EI V++
Sbjct: 255 AKM--SKNEEDI-----TSSLDGLTEKEIGVLN 332
>BG585420 similar to GP|17104707|gb unknown protein {Arabidopsis thaliana},
partial (37%)
Length = 784
Score = 26.2 bits (56), Expect = 4.7
Identities = 9/20 (45%), Positives = 14/20 (70%)
Frame = -2
Query: 4 RRVFTLSQVAQHKSNKDCWV 23
R V T+++ ++ NKDCWV
Sbjct: 759 RSVRTITKYQYYRDNKDCWV 700
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.136 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,534,890
Number of Sequences: 36976
Number of extensions: 34489
Number of successful extensions: 179
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 177
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 178
length of query: 143
length of database: 9,014,727
effective HSP length: 87
effective length of query: 56
effective length of database: 5,797,815
effective search space: 324677640
effective search space used: 324677640
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 53 (25.0 bits)
Lotus: description of TM0050.12


