

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
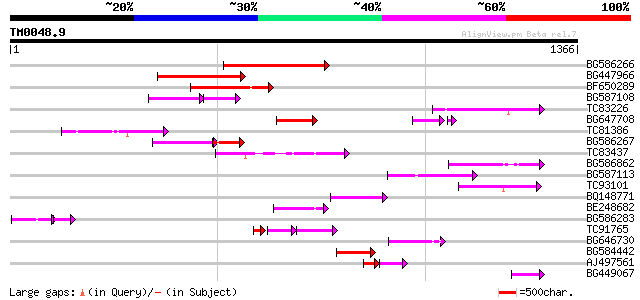
Query= TM0048.9
(1366 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-li... 223 3e-58
BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [impo... 223 4e-58
BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgari... 158 1e-38
BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - ... 90 4e-33
TC83226 weakly similar to PIR|G86419|G86419 probable reverse tra... 137 3e-32
BG647708 weakly similar to GP|13786450|gb| putative reverse tran... 120 3e-27
TC81386 similar to GP|10140689|gb|AAG13524.1 putative non-LTR re... 116 5e-26
BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At... 114 3e-25
TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imp... 110 3e-24
BG586862 103 3e-22
BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At... 95 2e-19
TC93101 90 5e-18
BQ148771 84 3e-16
BE248682 similar to GP|18568269|gb putative gag-pol polyprotein ... 84 4e-16
BG586283 similar to GP|7267666|em RNA-directed DNA polymerase-li... 54 9e-13
TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative ret... 71 3e-12
BG646730 70 6e-12
BG584442 68 2e-11
AJ497561 weakly similar to GP|10140689|gb putative non-LTR retro... 55 2e-10
BG449067 56 8e-08
>BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 789
Score = 223 bits (569), Expect = 3e-58
Identities = 110/256 (42%), Positives = 164/256 (63%)
Frame = -3
Query: 515 NQYRPISLCNVIFKIISKTIANRMKLILHDLISEAQSAFVPGRLITDNALIAFECFHYMK 574
++YR I+ CN +KII+K ++ RM+ +L +IS +QSAFVPGR I+DN LI + HY++
Sbjct: 775 SEYRTIAPCNTQYKIIAKILSKRMQPLLRSIISPSQSAFVPGRAISDNVLITHKILHYLR 596
Query: 575 KRISGRSGMMALKLDMSKAYDRVEWPFLRSVLTHMGFPVHWVNLVMNCVTTVNFAVMLNG 634
+ + + MA+K DM+KAYDR+ W FLR VLT +GF W++ +M CV+TV+++ ++NG
Sbjct: 595 QSGAKKHVSMAVKTDMTKAYDRIAWNFLREVLTRLGFHGIWISWIMECVSTVSYSFLING 416
Query: 635 NPQPTFAPHRGLRQGDPLSPYLFILCGEAFSALIQKSISTSALHGIKISRSAPVISHLLF 694
PQ P RGLRQGDPLSPYLFILC E S L Q+++ L G+K++R+ P I+HLLF
Sbjct: 415 GPQGRVLPSRGLRQGDPLSPYLFILCTEVLSGLCQQALRKGTLPGVKVARNCPPINHLLF 236
Query: 695 ADDSVLFARATVQEAECIKNILATYERASGQKINLEKSMLSVSRNVPENCFVDLQLLLGV 754
ADD++ F ++ + +I+ Y ASG+ IN KS ++ S + ++ L +
Sbjct: 235 ADDTMFFGKSNASSCAILLSIMDKYRAASGRCIN*TKSAITFSSKTSQAIIDRVKGELKI 56
Query: 755 KAVDSYDKYLGLPTII 770
KYLG I+
Sbjct: 55 AKEGGTGKYLGYRNIL 8
>BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [imported] -
Arabidopsis thaliana, partial (7%)
Length = 687
Score = 223 bits (568), Expect = 4e-58
Identities = 112/211 (53%), Positives = 146/211 (69%)
Frame = +2
Query: 357 WLKHGDRNTKFFHSKATQRRKRNLIEEIQDDNGRKFVRDADIARVLTSYFQEIFSTCNPT 416
WLK GD+NTKFFHSKA+QRRK N I++++D+ G + ++ R+L +YF +F++ NPT
Sbjct: 5 WLKDGDKNTKFFHSKASQRRKVNEIKKLKDETGNWCKGEENVERLLITYFNNLFTSSNPT 184
Query: 417 GIEEVATLVDGRVTDAHRRILSTPFTREDVEEALFQMHPTKAPGLDGFPALFYQKFWPIV 476
IEE +V G+++ H FT E+V EA+ QMHP KAPG DG PALF+QK+W IV
Sbjct: 185 AIEETCEVVKGKLSHEHIVWCEKEFTEEEVLEAINQMHPVKAPGPDGLPALFFQKYWHIV 364
Query: 477 GDDISEFCLQVLQGDISPGMVNQTLIVLIPKVKKAVFANQYRPISLCNVIFKIISKTIAN 536
G ++ + LQVL + +N+T IVLIPK K YRPISLCNV+ KII+K IAN
Sbjct: 365 GKEVQQMVLQVLNNSMETEELNKTFIVLIPKGKNPNTPKDYRPISLCNVVMKIITKVIAN 544
Query: 537 RMKLILHDLISEAQSAFVPGRLITDNALIAF 567
R+K L D+I QSAFV GRLITDNALIA+
Sbjct: 545 RVKQTLPDVIDVEQSAFVQGRLITDNALIAW 637
>BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgaris}, partial
(69%)
Length = 616
Score = 158 bits (400), Expect = 1e-38
Identities = 79/201 (39%), Positives = 125/201 (61%), Gaps = 1/201 (0%)
Frame = +3
Query: 436 ILSTPFTREDVEEALFQMHPTKAPGLDGFPALFYQKFWPIVGDDISEFCLQVLQGDISPG 495
+L + FT +V+ ALF M +KAPG+DG+ F++ W I+GD + + L + P
Sbjct: 9 LLCSEFTAVEVKNALFSMDSSKAPGIDGYNVHFFKCSWNIIGDSVIDAILDFFKTGFMPK 188
Query: 496 MVNQTLIVLIPKVKKAVFANQYRPISLCNVIFKIISKTIANRMKLILHDLISEAQSAFVP 555
++N T + L+PK +RPI+ C+VI+KIISK + +RM+ +L+ ++SE QSAFV
Sbjct: 189 IINCTYVTLLPKEVNVTSVKNFRPIACCSVIYKIISKILTSRMQGVLNSVVSENQSAFVK 368
Query: 556 GRLITDNALIAFECF-HYMKKRISGRSGMMALKLDMSKAYDRVEWPFLRSVLTHMGFPVH 614
GR+I DN +++ E Y +K IS R +K+D+ KAYD EWPF++ ++ +GFP
Sbjct: 369 GRVIFDNIILSHELVKSYSRKGISPR---CMVKIDLXKAYDSXEWPFIKHLMLELGFPYK 539
Query: 615 WVNLVMNCVTTVNFAVMLNGN 635
+VN VM +TT ++ NG+
Sbjct: 540 FVNWVMAXLTTASYTFNXNGD 602
>BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - Arabidopsis
thaliana, partial (17%)
Length = 677
Score = 90.1 bits (222), Expect(2) = 4e-33
Identities = 47/135 (34%), Positives = 66/135 (48%)
Frame = +2
Query: 335 EKNLDVLLKQEEIVWSQRSRANWLKHGDRNTKFFHSKATQRRKRNLIEEIQDDNGRKFVR 394
++ L K EE W Q+SR W GD N KF+H+ QR RN I + D +G
Sbjct: 11 KEKLQEAYKDEEDYWQQKSRNMWHISGDLNKKFYHALTKQRHARNRIVGLYDYDGNWITE 190
Query: 395 DADIARVLTSYFQEIFSTCNPTGIEEVATLVDGRVTDAHRRILSTPFTREDVEEALFQMH 454
+ + +V YF+++F PTG + + +T + L T E+V ALF MH
Sbjct: 191 EQGVEKVAVDYFEDLFQRTTPTGFDGFLDEITSSITPQMNQRLLRLATEEEVRLALFIMH 370
Query: 455 PTKAPGLDGFPALFY 469
P KAPG DG L +
Sbjct: 371 PEKAPGPDGMTTLLF 415
Score = 71.2 bits (173), Expect(2) = 4e-33
Identities = 40/89 (44%), Positives = 51/89 (56%)
Frame = +3
Query: 468 FYQKFWPIVGDDISEFCLQVLQGDISPGMVNQTLIVLIPKVKKAVFANQYRPISLCNVIF 527
F+Q W I+ D+ + L +N T I LIPK K+ + RPISLCNV +
Sbjct: 411 FFQHSWHIIKMDLLKMVNSFLASGKLDTRLNTTNICLIPKKKRPTRMTELRPISLCNVGY 590
Query: 528 KIISKTIANRMKLILHDLISEAQSAFVPG 556
KIISK + R+K+ L LISE QSAFV G
Sbjct: 591 KIISKVLCQRLKVCLPSLISETQSAFVHG 677
>TC83226 weakly similar to PIR|G86419|G86419 probable reverse transcriptase
100033-105622 [imported] - Arabidopsis thaliana, partial
(2%)
Length = 885
Score = 137 bits (345), Expect = 3e-32
Identities = 86/286 (30%), Positives = 138/286 (48%), Gaps = 15/286 (5%)
Frame = +3
Query: 1018 WLGTADGLYSVKTGYEFLQEVVAQDSPSSSLSRGLDQTL-WKRFWKAPSMPRVREVSWRV 1076
W+ G+YSVK+GY L+ Q ++S S D+TL WK+ W ++PR + + WR+
Sbjct: 6 WMHNPTGIYSVKSGYNTLRTWQTQQINNTSTSS--DETLIWKKIWSLHTIPRHKVLLWRI 179
Query: 1077 CTGALPVRTKLRQRGVDVDSSCPLCGLEDETVDHLFLGCNITRGCWFASPMGVRVD--PS 1134
+LPVR+ LR+RG+ CP C + ET+ HLF+ C +++ WF S + + D P+
Sbjct: 180 LNDSLPVRSSLRKRGIQCYPLCPRCHSKTETITHLFMSCPLSKRVWFGSNLCINFDNLPN 359
Query: 1135 LKMVDFLTMVIRDMDLEVVACVQRFLFAIWEARNKHIFEEKLFCIAGVLDRAASLVSQST 1194
++ L I D + + ++ +W ARN + E++ ++ RA++ +S
Sbjct: 360 PNFIN*LYEAIL*KDECITI*IAAIIYNLWHARNLSVLEDQTILEMDIIQRASNCISDYK 539
Query: 1195 LPFTLA------------ASQEKDNLKRWRRPAENVVKVNVDAAVGQDRFAGFGLVARDH 1242
T A + +W+RP +VKVN DA + G G++ RD
Sbjct: 540 QANTQAPPSMARTGYDPRSQHRPAKNTKWKRPNLGLVKVNTDANLQNHGKWGLGIIIRDE 719
Query: 1243 AGEVLAAAAKYPIMVLSPTVAEALSLRWAMDLAIQLGFRRVQFETD 1288
G V+AA+ AEA +L M A GF +V FE D
Sbjct: 720 VGLVMAASTWETDGNDRALEAEAYALLTGMRFAKDCGFXKVXFEGD 857
>BG647708 weakly similar to GP|13786450|gb| putative reverse transcriptase
{Oryza sativa}, partial (9%)
Length = 708
Score = 120 bits (302), Expect = 3e-27
Identities = 58/100 (58%), Positives = 75/100 (75%)
Frame = +1
Query: 642 PHRGLRQGDPLSPYLFILCGEAFSALIQKSISTSALHGIKISRSAPVISHLLFADDSVLF 701
P +GLRQGDPLSPYLFILC S L+++ + LHGI+++RS P I+HLLFADDS+LF
Sbjct: 4 PEKGLRQGDPLSPYLFILCANVLSGLLKREGNKQNLHGIQVARSDPKITHLLFADDSLLF 183
Query: 702 ARATVQEAECIKNILATYERASGQKINLEKSMLSVSRNVP 741
ARA + EA I +L +Y+ ASGQ +N EKS +S S+NVP
Sbjct: 184 ARANLTEAATIMQVLHSYQSASGQLVNFEKSEVSYSQNVP 303
Score = 40.0 bits (92), Expect(2) = 2e-04
Identities = 24/79 (30%), Positives = 40/79 (50%), Gaps = 1/79 (1%)
Frame = +3
Query: 970 GLTKVADLMLSGNRGWNVPLIEWTFCPATASRIMSVPLPRQPESDQLFWLGTADGLYSVK 1029
G V +L+ + WN LI +F A +I+ +PL + D++ W DG+YSV+
Sbjct: 390 GSLSVDELIDYDTKQWNRDLIFHSFNNYAAHQIIKIPLSMRQPEDKIIWHWEKDGIYSVR 569
Query: 1030 TGYEFL-QEVVAQDSPSSS 1047
+ + L E + + SSS
Sbjct: 570 SAHHALCDENFSNQAESSS 626
Score = 24.3 bits (51), Expect(2) = 2e-04
Identities = 7/21 (33%), Positives = 11/21 (52%)
Frame = +2
Query: 1056 LWKRFWKAPSMPRVREVSWRV 1076
+WK W P+ V+ WR+
Sbjct: 641 IWKAIWNTPASRSVQNFLWRL 703
>TC81386 similar to GP|10140689|gb|AAG13524.1 putative non-LTR retroelement
reverse transcriptase {Oryza sativa (japonica
cultivar-group)}, partial (1%)
Length = 798
Score = 116 bits (291), Expect = 5e-26
Identities = 74/267 (27%), Positives = 130/267 (47%), Gaps = 11/267 (4%)
Frame = -1
Query: 126 WEMIRSVRPMAPIPWLCIGDFNDILSPSDKLGGAPPDLARLQVAT-QACADCG-LQRVDF 183
W I +++ + W CIGDFN IL + G P ARL + Q +D L +
Sbjct: 783 WSAISNIQTQHKLSWCCIGDFNTILGSHEHQGSHTP--ARLPMLDFQQWSDVNNLFHLPT 610
Query: 184 SGPSFTWTNNRVGPGRVDERLDYALINQAWVNLWPVSSVSHLIRHQSDHNPVVLHCGSRR 243
G +FTWTN R G +RLD +++NQ ++ S L + +SDH P++ ++
Sbjct: 609 RGSAFTWTNGRRGRNNTRKRLDRSIVNQLMIDNCESLSACTLTKLRSDHFPILFELQTQN 430
Query: 244 TETQRHRTRMFRFEEVWLESGEECAEIISEGWSNTNLS-----ILSRINLVGRSLDSWGR 298
+ + F+F ++W + +C II + W+N + + ++ + L W +
Sbjct: 429 IQF----SSSFKFMKMW-SAHPDCINIIKQCWANQVVGCPMFVLNQKLKNLKEVLKVWNK 265
Query: 299 EKYGELPKRIKEARAFLQRLQEEVQT----EQVVRATREAEKNLDVLLKQEEIVWSQRSR 354
+G + ++ A L +Q ++ + + ++ + A+ NL+ L EE+ W ++S+
Sbjct: 264 NTFGNVHSQVDNAYKELDDIQVKIDSIGYSDVLMDQEKAAQLNLESALNIEEVFWHEKSK 85
Query: 355 ANWLKHGDRNTKFFHSKATQRRKRNLI 381
NW GDRNT FFH A +R +LI
Sbjct: 84 VNWHCEGDRNTAFFHRVAKIKRTSSLI 4
>BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (16%)
Length = 794
Score = 114 bits (284), Expect = 3e-25
Identities = 59/159 (37%), Positives = 94/159 (59%), Gaps = 1/159 (0%)
Frame = +3
Query: 345 EEIVWSQRSRANWLKHGDRNTKFFHSKATQRRKRNLIEEIQDDNGRKFVRDADIARVLTS 404
EEI W Q+SR NWL+ GDRNTKFFH+ RR +N I + DD+ +++ + D+ R+ S
Sbjct: 123 EEIFWMQKSRLNWLRSGDRNTKFFHAVTKNRRAQNRILSLIDDDDKEWFVEEDLGRLADS 302
Query: 405 YFQEIFSTCNPTGIEEVATLVDGRVTDAHRRILSTPFTREDVEEALFQMHPTKAPGLDGF 464
+F+ ++S+ + E + VT+ L +RE+V EA+F ++P K PG DG
Sbjct: 303 HFKLLYSSEDVGITLEDWNSIPAIVTEEQNAQLMAQISREEVREAVFDINPHKCPGPDGM 482
Query: 465 PALFYQKFWPIVGDDISEFCLQVLQ-GDISPGMVNQTLI 502
F+Q+FW +GDD++ + L+ G + G +N+T I
Sbjct: 483 NVFFFQQFWDTMGDDLTSMAQEFLRTGKLEEG-INKTNI 596
Score = 63.2 bits (152), Expect = 7e-10
Identities = 35/78 (44%), Positives = 52/78 (65%)
Frame = +1
Query: 489 QGDISPGMVNQTLIVLIPKVKKAVFANQYRPISLCNVIFKIISKTIANRMKLILHDLISE 548
QG++ QT L+PK +A ++RPISLCNV +KI+SK ++ R+K +L +I+E
Sbjct: 559 QGNLKRESTKQTS-GLVPKKLEAKRLVEFRPISLCNVAYKIVSKVLSKRLKSVLPWIITE 735
Query: 549 AQSAFVPGRLITDNALIA 566
Q+AF +LI+DN LIA
Sbjct: 736 TQAAFGRRQLISDNILIA 789
>TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imported] -
Arabidopsis thaliana, partial (6%)
Length = 951
Score = 110 bits (276), Expect = 3e-24
Identities = 89/337 (26%), Positives = 145/337 (42%), Gaps = 15/337 (4%)
Frame = +2
Query: 497 VNQTLIVLIPKVKKAVFANQYRPISLCNVIFKIISKTIANRMKLILHDLISEAQSAFVPG 556
+N T I LIPKV N +RPISL ++KI+ K +ANR+++++ +IS+AQSAFV
Sbjct: 50 INSTFIALIPKVDNPQRLNDFRPISLVGSLYKILGKLLANRLRVVIGSVISDAQSAFVKN 229
Query: 557 RLITDNALI--------------AFECFHYMKKRISGRS-GMMALKLDMSKAYDRVEWPF 601
R I + + F C ++ KR+ S G++ + +
Sbjct: 230 RQILEMVFL*QMRLWMRLRN*RKIFCCLRWILKRLITLSIGLIWILF*VG---------- 379
Query: 602 LRSVLTHMGFPVHWVNLVMNCVTTVNFAVMLNGNPQPTFAPHRGLRQGDPLSPYLFILCG 661
M F V W + CV+T +V++NG+P
Sbjct: 380 -------MSFLVLWRKWIKECVSTATTSVLVNGSPTNVL--------------------- 475
Query: 662 EAFSALIQKSISTSALHGIKISRSAPVISHLLFADDSVLFARATVQEAECIKNILATYER 721
+L+Q + T G+ + V+SHL FA+D++L ++ L +
Sbjct: 476 --MKSLVQTQLFTRYSFGVV---NPVVVSHLQFANDTLLLETKNWANIRALRAALVIF*A 640
Query: 722 ASGQKINLEKSMLSVSRNVPENCFVDLQLLLGVKAVDSYDKYLGLPTIIGKSKTQIFRFV 781
SG K+N KS L V N+ + + +L K YLG+P + + +
Sbjct: 641 MSGLKVNFHKSGL-VCVNIAPSWLSEAASVLSWKVGKVPFLYLGMPIEGNSRRLSFWEPI 817
Query: 782 KERVWKKLKGWKEQTLSRAGREVLIKSVAQAIPSYVM 818
R+ +L GW + LS GR VL+KSV ++ Y +
Sbjct: 818 VNRIKARLTGWNSRFLSFGGRLVLLKSVLTSLSVYAL 928
>BG586862
Length = 804
Score = 103 bits (258), Expect = 3e-22
Identities = 72/233 (30%), Positives = 113/233 (47%), Gaps = 2/233 (0%)
Frame = -1
Query: 1058 KRFWKAPSMPRVREVSWRVCTGALPVRTKLRQRGVDVDSSCPLCGLEDETVDHLFLGCNI 1117
++ W ++PR + WR+ ALPV+ +L +RG+ CP C + ETV HLFL C +
Sbjct: 660 EKVWGIKTIPRHKSFLWRLLHNALPVKDELHKRGIRCSLLCPRCESKIETVQHLFLNCEV 481
Query: 1118 TRGCWFASPMGVRVDPS--LKMVDFLTMVIRDMDLEVVACVQRFLFAIWEARNKHIFEEK 1175
T+ WF S +G+ S L D++T I D E + + L++IW ARN+ +FE
Sbjct: 480 TQKEWFGSQLGINFHSSGVLHFHDWITNFILKNDEETIIALTALLYSIWHARNQKVFENI 301
Query: 1176 LFCIAGVLDRAASLVSQSTLPFTLAASQEKDNLKRWRRPAENVVKVNVDAAVGQDRFAGF 1235
V+ RA+S S F +A + +++V+ N A+ G
Sbjct: 300 DVPGDVVIQRASS----SLHSFKMA------------QVSDSVLPSN---AIPSYSLWGI 178
Query: 1236 GLVARDHAGEVLAAAAKYPIMVLSPTVAEALSLRWAMDLAIQLGFRRVQFETD 1288
G+VAR+ G +A+ + T AEA + AM A GF + +FE+D
Sbjct: 177 GVVARNCEGLAMASGTWLRHGIPCATTAEAWGIYQAMVFAGDCGFSKFEFESD 19
>BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (10%)
Length = 767
Score = 94.7 bits (234), Expect = 2e-19
Identities = 66/227 (29%), Positives = 98/227 (43%), Gaps = 10/227 (4%)
Frame = -3
Query: 911 AKKGYRPSYAWSRILKTSAMIQKGSCWRIGEGSRVRIWEDN--W----LANGPPVNFRQ- 963
A G SYAW I +I++G+ IG G IWE W + N P + +
Sbjct: 756 APLGSWASYAWRSIHSAQHLIKQGAKVIIGNGENTNIWEREMAWKLTCVTNHPNKHSSRA 577
Query: 964 ---DVVDELGLTKVADLMLSGNRGWNVPLIEWTFCPATASRIMSVPLPRQPESDQLFWLG 1020
+ + D M R N LI F T +I+S+ D W
Sbjct: 576 Y*APTLYGYEGCRSDDPM---RRERNANLINSIFPEGTRRKILSIHPQGPIGEDSYSWEY 406
Query: 1021 TADGLYSVKTGYEFLQEVVAQDSPSSSLSRGLDQTLWKRFWKAPSMPRVREVSWRVCTGA 1080
+ G YSVK+GY ++A + ++ + L++R WK + P+VR WR + +
Sbjct: 405 SKSGHYSVKSGYYVQTNIIAAANQRGTVDQPSLDDLYQRVWKYNTSPKVRHFLWRCISNS 226
Query: 1081 LPVRTKLRQRGVDVDSSCPLCGLEDETVDHLFLGCNITRGCWFASPM 1127
LP +R R + D SC CG+E ETV+H+ C R W SP+
Sbjct: 225 LPTAANMRSRHISKDGSCSRCGMESETVNHILFQCPYARLIWATSPI 85
>TC93101
Length = 675
Score = 90.1 bits (222), Expect = 5e-18
Identities = 61/221 (27%), Positives = 96/221 (42%), Gaps = 20/221 (9%)
Frame = -1
Query: 1081 LPVRTKLRQRGVDVDSSCPLCGLEDETVDHLFLGCNITRGCWFASPMGVRV--DPSLKMV 1138
LPVR +L +RGV+ CP C ET +H+F+ C T+ WF S + +R + ++
Sbjct: 663 LPVRXELNKRGVNCPPLCPRCYFNLETTNHIFMSCERTQRVWFGSQLSIRFPDNSTINFS 484
Query: 1139 DFLTMVIRDMDLEVVACVQRFLFAIWEARNKHIFEEKLFCIAGVLDRAAS---------- 1188
D+L I + E++ + ++IW ARNK IFE + ++ A +
Sbjct: 483 DWLFDAISNQTEEIIIKISAITYSIWHARNKAIFENQFVSEDTIIQ*AQNSILAYEQATI 304
Query: 1189 --------LVSQSTLPFTLAASQEKDNLKRWRRPAENVVKVNVDAAVGQDRFAGFGLVAR 1240
L S S T + + RW++P N++K N DA + G G + R
Sbjct: 303 KPQNPNIVLSSLSATSNTNTTRRRSNVRSRWQKPLNNILKANCDANLQVQGRWGLGCIIR 124
Query: 1241 DHAGEVLAAAAKYPIMVLSPTVAEALSLRWAMDLAIQLGFR 1281
+ GE A AE ++ AM LA GF+
Sbjct: 123 NADGEAKVTATWCINGFDCAATAETYAILAAMYLAKDYGFK 1
>BQ148771
Length = 680
Score = 84.3 bits (207), Expect = 3e-16
Identities = 50/139 (35%), Positives = 69/139 (48%), Gaps = 1/139 (0%)
Frame = -3
Query: 772 KSKTQIFRF-VKERVWKKLKGWKEQTLSRAGREVLIKSVAQAIPSYVMSCFVLPDGLCNE 830
+ T+ RF V +V L WK LS A R L KSV +A+P Y M ++P E
Sbjct: 627 RKSTKKSRFSVYYQVHVMLANWKANHLSLARRVTLAKSVIEAVPLYPMMTTIIPKACIEE 448
Query: 831 IESMISRFYWGGDASKRGIHWVKWKTMCQPKHVGGLGFRDFKSFNLALVAKNWWRIYNYP 890
I+ + +F WG R H V W+TM +PK + GLG R N A + K W IY+
Sbjct: 447 IQKLQRKFVWGDTEVSRRYHAVGWETMSKPKTIYGLGLRRLDVMNKACIMKLGWSIYSGS 268
Query: 891 DSLLGKVFKSVYFSSGRME 909
+SL +V + Y S +E
Sbjct: 267 NSLCTEVMRGKYQRSESLE 211
>BE248682 similar to GP|18568269|gb putative gag-pol polyprotein {Zea mays},
partial (1%)
Length = 441
Score = 84.0 bits (206), Expect = 4e-16
Identities = 46/131 (35%), Positives = 69/131 (52%)
Frame = +3
Query: 637 QPTFAPHRGLRQGDPLSPYLFILCGEAFSALIQKSISTSALHGIKISRSAPVISHLLFAD 696
Q + RGL+QGDPL+P+LF+L E S L++ +++ + G + R +SHL +AD
Sbjct: 3 QEEISVQRGLKQGDPLAPFLFLLVAEGISGLMKNAVNRNLFQGFDVKRGGTRVSHLQYAD 182
Query: 697 DSVLFARATVQEAECIKNILATYERASGQKINLEKSMLSVSRNVPENCFVDLQLLLGVKA 756
D++ TV +K +L +E ASG K+N KS L + NVP + L +
Sbjct: 183 DTLCIGMPTVDNLWTLKALLQGFEMASGLKVNFHKSSL-IGINVPRDFMEAACRFLNCRE 359
Query: 757 VDSYDKYLGLP 767
YLGLP
Sbjct: 360 ESIPFIYLGLP 392
>BG586283 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (7%)
Length = 774
Score = 53.5 bits (127), Expect(2) = 9e-13
Identities = 35/108 (32%), Positives = 56/108 (51%)
Frame = +2
Query: 4 TSMRLLSWNCRGLGNLEAVRALRGLIHSQAPDIVFLMETKLFSFEMQRHRGMGGLSNIFP 63
+S+ + WNC+G+G+ VR LR +PDI+FLMETK ++ + +N F
Sbjct: 197 SSLVNMCWNCQGIGSDLTVRRLREFQTKDSPDIMFLMETKRKDEDVYKMYRGTEFTNHFT 376
Query: 64 VQCGRNRAGGLCLLWRSEVEVTIINASLHHILFTVVHSDSVTVPMQVY 111
V +GGL L W+ V+V I+ +S + I V ++D +Y
Sbjct: 377 VP-PVGLSGGLALSWKDNVQVEILASSANVIDTKVNYNDKSFFVSYIY 517
Score = 39.3 bits (90), Expect(2) = 9e-13
Identities = 16/52 (30%), Positives = 27/52 (51%)
Frame = +3
Query: 107 PMQVYALYGFPELHLKDRTWEMIRSVRPMAPIPWLCIGDFNDILSPSDKLGG 158
P + G P+ + + W+ + ++ WL GDFND+L S+K+GG
Sbjct: 495 PFSYHTSIGVPQPEHRAKFWDELSTIGAQRDGAWLLTGDFNDLLDNSEKVGG 650
>TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative retroelement
{Oryza sativa (japonica cultivar-group)}, partial (1%)
Length = 625
Score = 71.2 bits (173), Expect = 3e-12
Identities = 40/99 (40%), Positives = 57/99 (57%)
Frame = +3
Query: 692 LLFADDSVLFARATVQEAECIKNILATYERASGQKINLEKSMLSVSRNVPENCFVDLQLL 751
LLF + R A+ +KNIL YE SG+ I+L KS + SRNVP+ + +
Sbjct: 279 LLFEGAPLRSLRVEEHHAQIMKNILILYEEDSGKAISLRKS*IYCSRNVPDILKTSITYI 458
Query: 752 LGVKAVDSYDKYLGLPTIIGKSKTQIFRFVKERVWKKLK 790
LGV+ + KYLGLP++IG+ +T F +K VW+K K
Sbjct: 459 LGVQFMLGTCKYLGLPSMIGRDRTTTFSSIKGGVWQKNK 575
Score = 58.5 bits (140), Expect(2) = 3e-11
Identities = 29/69 (42%), Positives = 40/69 (57%)
Frame = +2
Query: 622 CVTTVNFAVMLNGNPQPTFAPHRGLRQGDPLSPYLFILCGEAFSALIQKSISTSALHGIK 681
CV + ++ V++N + P RGL+QGD LSPY+FI+C E S LI + HG
Sbjct: 104 CVESNDYYVLVNNDAVDPIIPSRGLQQGDHLSPYIFIICVEGLSFLIPHAKERGDTHGTS 283
Query: 682 ISRSAPVIS 690
I R AP +S
Sbjct: 284 I*RGAPPVS 310
Score = 28.9 bits (63), Expect(2) = 3e-11
Identities = 11/29 (37%), Positives = 19/29 (64%)
Frame = +1
Query: 588 LDMSKAYDRVEWPFLRSVLTHMGFPVHWV 616
L +SK Y+RV+ +L+ ++ MGF W+
Sbjct: 1 LYISKVYNRVD*DYLKEIMIKMGFNNRWI 87
>BG646730
Length = 799
Score = 70.1 bits (170), Expect = 6e-12
Identities = 43/138 (31%), Positives = 69/138 (49%), Gaps = 2/138 (1%)
Frame = +1
Query: 914 GYRPSYAWSRILKTSAMIQKGSCWRIGEGSRVRIWEDNWLANGPPVNFRQDVVDELGLTK 973
G +PSYAW + +I GS W I G VRIW+D+WL N +VD T
Sbjct: 388 GNQPSYAWRSMFNIKDVIDLGSRWSISNGQNVRIWKDDWLPNQTGFKVCSPMVDFEEATC 567
Query: 974 VADLMLSGNRGWNVPLIEWTFCPATASRIMSVPLPRQ--PESDQLFWLGTADGLYSVKTG 1031
+++L+ + + L+ F A +I+++PL + P+ L+W D YSV++
Sbjct: 568 ISELIDTNTKSSKRDLVRNVFSAFEAKQILNIPLSWRLWPDRRILYW--ERDRNYSVRSA 741
Query: 1032 YEFLQEVVAQDSPSSSLS 1049
+ + +V +DSP S S
Sbjct: 742 HHLI-KVATKDSPEVSSS 792
>BG584442
Length = 775
Score = 68.2 bits (165), Expect = 2e-11
Identities = 35/96 (36%), Positives = 60/96 (62%), Gaps = 1/96 (1%)
Frame = +1
Query: 787 KKLKGWKEQTLSRAGREVLIKSVAQAIPSYVMSCFVLPDGLCNEIESMISRFYW-GGDAS 845
KK+ + + LS+ EV+IK Q+I SYVMS F+L + +EIE +++ F W +
Sbjct: 379 KKINF*RNKCLSKVM*EVMIKYALQSISSYVMSIFLLLNSQVDEIEKIMNTFSWVHVGEN 558
Query: 846 KRGIHWVKWKTMCQPKHVGGLGFRDFKSFNLALVAK 881
++G+HW+ + + K+ GG+GF DF +FN+ ++ K
Sbjct: 559 RKGMHWMS*EKLFVHKNYGGMGFTDFTTFNIPMLGK 666
>AJ497561 weakly similar to GP|10140689|gb putative non-LTR retroelement
reverse transcriptase {Oryza sativa (japonica
cultivar-group)}, partial (2%)
Length = 621
Score = 55.5 bits (132), Expect(2) = 2e-10
Identities = 23/66 (34%), Positives = 36/66 (53%)
Frame = +2
Query: 892 SLLGKVFKSVYFSSGRMECAKKGYRPSYAWSRILKTSAMIQKGSCWRIGEGSRVRIWEDN 951
SLL K+ K YF A G+ PS+ W +L T +++ G W IG+GS++ + +
Sbjct: 410 SLLSKILKFKYFPQWDFSYANLGHNPSFTWRSLLSTQSLLTLGHRWMIGDGSQINVSSMS 589
Query: 952 WLANGP 957
W+ N P
Sbjct: 590 WIRNRP 607
Score = 29.6 bits (65), Expect(2) = 2e-10
Identities = 11/38 (28%), Positives = 25/38 (64%)
Frame = +3
Query: 853 KWKTMCQPKHVGGLGFRDFKSFNLALVAKNWWRIYNYP 890
++ + + K VGGLG ++ + FN++++ K W++ + P
Sbjct: 300 EFNNLARHKSVGGLGLQN-QEFNISMLGKQRWKLLSQP 410
>BG449067
Length = 578
Score = 56.2 bits (134), Expect = 8e-08
Identities = 28/79 (35%), Positives = 46/79 (57%)
Frame = -2
Query: 1210 RWRRPAENVVKVNVDAAVGQDRFAGFGLVARDHAGEVLAAAAKYPIMVLSPTVAEALSLR 1269
+W++P ++++K+N DA + G G+VAR+ G +A+ + S T AEA +
Sbjct: 310 KWKKPEKDIIKLNSDANLSSTDLWGIGVVARNDEGFAMASGTWFRFGFPSATTAEAWGIY 131
Query: 1270 WAMDLAIQLGFRRVQFETD 1288
AM A + GF +VQFE+D
Sbjct: 130 QAMIFAGEYGFSKVQFESD 74
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.137 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 47,072,942
Number of Sequences: 36976
Number of extensions: 704563
Number of successful extensions: 3202
Number of sequences better than 10.0: 74
Number of HSP's better than 10.0 without gapping: 3127
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3177
length of query: 1366
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1258
effective length of database: 5,021,319
effective search space: 6316819302
effective search space used: 6316819302
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0048.9


