

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
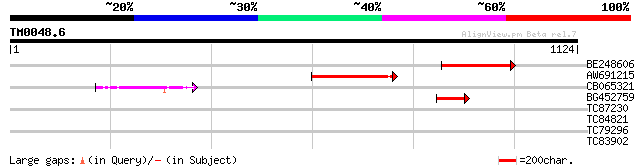
Query= TM0048.6
(1124 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BE248606 homologue to SP|P15001|PHY Phytochrome A. [Garden pea] ... 238 7e-63
AW691215 homologue to GP|6502525|gb phytochrome B {Pisum sativum... 144 2e-34
CB065321 similar to GP|17980438|gb bacteriophytochrome 1 {Pseudo... 102 8e-22
BG452759 similar to GP|6502525|gb| phytochrome B {Pisum sativum}... 62 1e-09
TC87230 similar to SP|Q06215|PPO_VICFA Polyphenol oxidase A1 ch... 31 2.4
TC84821 weakly similar to GP|17065212|gb|AAL32760.1 Unknown prot... 31 2.4
TC79296 weakly similar to GP|14279169|gb|AAK58515.1 beta-1 3-glu... 29 9.0
TC83902 similar to SP|O81974|C7D8_SOYBN Cytochrome P450 71D8 (EC... 29 9.0
>BE248606 homologue to SP|P15001|PHY Phytochrome A. [Garden pea] {Pisum
sativum}, partial (12%)
Length = 449
Score = 238 bits (608), Expect = 7e-63
Identities = 121/147 (82%), Positives = 134/147 (90%)
Frame = +1
Query: 856 KKLDVEGLVTGVFCFLQLASPELQQALHIQHLSEQTALKRLKALTYMKRQIRNPLSGIVF 915
+K+D EGLVTGVFCFLQLASPELQQALHIQ LSEQTALKRLK LTYM+RQIRNPLSGIVF
Sbjct: 7 EKIDAEGLVTGVFCFLQLASPELQQALHIQRLSEQTALKRLKVLTYMRRQIRNPLSGIVF 186
Query: 916 SRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDSDLDSIMDGYLDLEMAEFTLQDVLIT 975
S K LE T+LG EQKR+V+TS+QCQRQLSKILDDSDLDSI+DGYLDLEMAEFTL +VL+T
Sbjct: 187 SSKMLENTELGTEQKRIVNTSSQCQRQLSKILDDSDLDSIIDGYLDLEMAEFTLHEVLVT 366
Query: 976 SLSQIMARSSARGIRIVNDVAEEIMVE 1002
SLSQ+M RS+ R IRIVNDVAE I +E
Sbjct: 367 SLSQVMNRSNTRSIRIVNDVAEHIAME 447
>AW691215 homologue to GP|6502525|gb phytochrome B {Pisum sativum}, partial
(15%)
Length = 554
Score = 144 bits (363), Expect = 2e-34
Identities = 84/176 (47%), Positives = 118/176 (66%), Gaps = 5/176 (2%)
Frame = +2
Query: 599 IDTRLSDLKIEGMQELEAVTSEMVRLIETATVPILAVDIDGLVNGWNIKIAELTGLPVGE 658
+ T +++L+++G+ EL +V EMVRLIETAT PI AVD++G +NGWN K++ELTGL V +
Sbjct: 32 VHTHMAELELQGVDELSSVAREMVRLIETATAPIFAVDVNGRINGWNAKVSELTGLLVED 211
Query: 659 AIGKHLL-TLVEDCSTDRVKKMLDLALSGEEEKNVQFEIKTHGSKMESGPISLVVNACAS 717
A+GK LL LV S + V K+L AL GEE+KNV+ +++T G ++ + +VVNAC+S
Sbjct: 212 AMGKSLLHDLVYKESQETVDKLLSHALKGEEDKNVEIKMRTFGPGNQNKAVFIVVNACSS 391
Query: 718 RDLRE-NVVGVCFVAQDITAQKTVMDKF---TRIEGDYKAIVQNPNPLIPPIFGTD 769
+ L + V + QD+T QK VMDKF TR Y +PN LIPPIF +D
Sbjct: 392 KVLYK*YSWSVL*LVQDVTGQKVVMDKFH*HTR*LQGY--CT*DPNALIPPIFASD 553
Score = 29.6 bits (65), Expect = 6.9
Identities = 15/38 (39%), Positives = 21/38 (54%)
Frame = +2
Query: 764 PIFGTDEFGWCCEWNPAMTKLTGWKREEVMDKMLLGEV 801
PIF D G WN +++LTG E+ M K LL ++
Sbjct: 128 PIFAVDVNGRINGWNAKVSELTGLLVEDAMGKSLLHDL 241
>CB065321 similar to GP|17980438|gb bacteriophytochrome 1 {Pseudomonas
putida}, partial (29%)
Length = 659
Score = 102 bits (254), Expect = 8e-22
Identities = 65/207 (31%), Positives = 113/207 (54%), Gaps = 4/207 (1%)
Frame = +2
Query: 170 IIHRVTGSLIIDFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQE 229
+ HR LI +FEP + +T G Y L ++ L + ++E+L V++
Sbjct: 107 LAHRHDQVLIAEFEPSR-LGSELTEQG--DYYPLVRGFVSSLHH--AETLEQLLQQSVEQ 271
Query: 230 VFELTGYDRVMAYKFHEDDHGEVIAEITKPGLEPYLGLHYPATDIPQASRFLFMKNKVRM 289
+ +TG+ RV AY+F + +G+V+AE+ G YLGLH+PA+DIP+ +R L+ N++R+
Sbjct: 272 IRRITGFGRVKAYRFDAEGNGQVLAEVADAGYPRYLGLHFPASDIPRQARELYRINRIRV 451
Query: 290 IVDCHAKQVKVLIDEK----LPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVND 345
I D + + ++ P D++ + LR+ HLQYM NM ++AS+ ++++V
Sbjct: 452 IEDANYQPSPLVPGNNPRTGKPLDMSF--AALRSVSPVHLQYMRNMGTLASMSLSIMV-- 619
Query: 346 NDEDGDGSDSVQPQKRKRLWGLVVCHN 372
DG+ LWGL++CH+
Sbjct: 620 ---DGE------------LWGLILCHH 655
>BG452759 similar to GP|6502525|gb| phytochrome B {Pisum sativum}, partial
(7%)
Length = 676
Score = 62.4 bits (150), Expect = 1e-09
Identities = 25/65 (38%), Positives = 44/65 (67%)
Frame = +1
Query: 846 KYVECLLSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQHLSEQTALKRLKALTYMKRQ 905
K+V L+ + +++++G + G FCFLQ+ SPELQQAL + + + R+K L Y+ ++
Sbjct: 403 KFVXTFLTANXRVNMDGQIIGAFCFLQIVSPELQQALTVXRQQDSSCFARMKXLXYICQE 582
Query: 906 IRNPL 910
++NPL
Sbjct: 583 VKNPL 597
>TC87230 similar to SP|Q06215|PPO_VICFA Polyphenol oxidase A1 chloroplast
precursor (EC 1.10.3.1) (PPO) (Catechol oxidase). [Broad
bean], partial (79%)
Length = 1778
Score = 31.2 bits (69), Expect = 2.4
Identities = 15/45 (33%), Positives = 22/45 (48%)
Frame = +3
Query: 307 PFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNDNDEDGD 351
PF+ GS + PHS H Q M N++S + ++ D D D
Sbjct: 1644 PFNTEFAGSFVNVPHSLHEQTMKNINSSCRFGLTDLLGDLGVDDD 1778
>TC84821 weakly similar to GP|17065212|gb|AAL32760.1 Unknown protein
{Arabidopsis thaliana}, partial (31%)
Length = 534
Score = 31.2 bits (69), Expect = 2.4
Identities = 15/33 (45%), Positives = 23/33 (69%), Gaps = 1/33 (3%)
Frame = -2
Query: 139 SALQKALGFAEVTL-LNPILVHCKTSGKPFYAI 170
++L+K LGFA + L +NP+ C+TS K F+ I
Sbjct: 146 NSLKKNLGFACINLFINPVSPSCQTSSKSFHTI 48
>TC79296 weakly similar to GP|14279169|gb|AAK58515.1 beta-1 3-glucanase-like
protein {Olea europaea}, partial (90%)
Length = 1710
Score = 29.3 bits (64), Expect = 9.0
Identities = 18/39 (46%), Positives = 22/39 (56%)
Frame = -3
Query: 860 VEGLVTGVFCFLQLASPELQQALHIQHLSEQTALKRLKA 898
V GLV G FC+L LA P I + E++ LKR KA
Sbjct: 1183 VTGLVDGGFCWLDLAKP-------ISYDMERSVLKRPKA 1088
>TC83902 similar to SP|O81974|C7D8_SOYBN Cytochrome P450 71D8 (EC 1.14.-.-)
(P450 CP7). [Soybean] {Glycine max}, partial (41%)
Length = 900
Score = 29.3 bits (64), Expect = 9.0
Identities = 17/51 (33%), Positives = 27/51 (52%)
Frame = -2
Query: 973 LITSLSQIMARSSARGIRIVNDVAEEIMVEILYGDSLRLQQVLADFLLISI 1023
L SLS A S G I++D+A I+V+++ + Q+ A LL+ I
Sbjct: 329 LNASLSSACAFSLTLGFFIISDIAHSIVVDVVSVPATNTSQITALMLLVVI 177
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.135 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,197,786
Number of Sequences: 36976
Number of extensions: 399568
Number of successful extensions: 1919
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 1897
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1915
length of query: 1124
length of database: 9,014,727
effective HSP length: 106
effective length of query: 1018
effective length of database: 5,095,271
effective search space: 5186985878
effective search space used: 5186985878
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0048.6


