

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
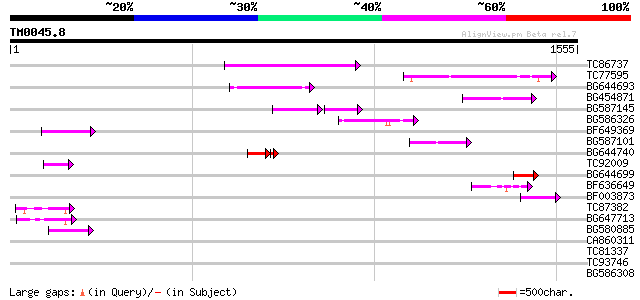
Query= TM0045.8
(1555 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 258 2e-68
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 182 7e-46
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 162 1e-39
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 121 2e-27
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 92 3e-27
BG586326 similar to PIR|G84493|G8 probable retroelement pol poly... 103 4e-22
BF649369 88 3e-17
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 78 3e-14
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 61 4e-12
TC92009 60 9e-09
BG644699 similar to PIR|T07863|T078 probable polyprotein - pinea... 59 1e-08
BF636649 similar to GP|21628724|emb OSJNBa0033H08.7 {Oryza sativ... 56 1e-07
BF003873 similar to GP|14715222|em putative polyprotein {Cicer a... 53 8e-07
TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulch... 52 2e-06
BG647713 homologue to GP|15042313|gb| 232R {Chilo iridescent vir... 48 3e-05
BG580885 43 8e-04
CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product ... 42 0.002
TC81337 similar to GP|15723293|gb|AAL06332.1 U2 auxiliary factor... 39 0.016
TC93746 weakly similar to GP|22830935|dbj|BAC15800. hypothetical... 39 0.021
BG586308 weakly similar to PIR|F84528|F8 probable retroelement p... 36 0.10
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 258 bits (658), Expect = 2e-68
Identities = 150/390 (38%), Positives = 228/390 (58%), Gaps = 16/390 (4%)
Frame = +1
Query: 589 LLTEFPEVFQEPK--ELPPKRAT-DHAILL---QEG--APIPNIRPYRYPFYQKNEIEKL 640
+L EFP++F K ++P R DHAI L ++G P+P Y + ++K
Sbjct: 343 VLEEFPDLFNPEKAYQVPASRGLLDHAIPLIPDKDGNDPPLPWGPLYGMSRQELLVLKKT 522
Query: 641 VKEMLAAGIIRHSTSPFSSPAILVKKKDGGWRFCVDYRALNKVTIPDKFPIPIIDELLDE 700
++++L G I+ S S +P + V+K GG RFCVDYRALN +T D++P+P+I E L
Sbjct: 523 LEDLLDKGFIKASGSAAGAPVLFVRKPGGGIRFCVDYRALNAITKKDRYPLPLISETLRR 702
Query: 701 IGTAEIFSKLDLKSGYHQIRMREEDIPKTAFRTHEGHYEYLVLPFGLTNAPSTFQALMNQ 760
+ A F+KLD+ + +H++R+++ED KTAFRT G +E++V PFGLT AP+TFQ +N+
Sbjct: 703 VAGARWFTKLDVVAAFHKMRIKDEDQEKTAFRTRYGLFEWIVCPFGLTGAPATFQRYINK 882
Query: 761 VLRPYLRKFVLVFFYDILIYSNNVDL-HKEHLREVLQVLRDNHLVANQKKCSFGQSELIY 819
L +L FV + D+LIY+ H+ +R VL+ L D L + KKC F + + Y
Sbjct: 883 TLHEFLDDFVTAYIDDVLIYTTGSKKDHEAQVRRVLRRLADAGLSLDPKKCEFSVTTVKY 1062
Query: 820 LGHVISK-EGVAADPSKIKDMLNWPLPKDVKGLRGFLGLTGYYRRFVRNYSKLAQPLNQL 878
+G +++ +GV+ DP K+ + +W P VKG R FLG YY+ F+ YS++ +PL +L
Sbjct: 1063VGFILTAGKGVSCDPLKLAAIRDWLPPGSVKGARSFLGFCNYYKDFIPGYSEITEPLTRL 1242
Query: 879 LKKN-NFSWSAGATQAFDKLKEIMTTVPVLAVPDFQKTFVLETDASGKGLGAVLMQ---- 933
+K+ F W A AF KLK + PVL + D + +ETD SG LG VL Q
Sbjct: 1243TRKDFPFRWGAEQEAAFTKLKRLFAEEPVLRMFDPEAVTTVETDCSGFALGGVLTQEDGT 1422
Query: 934 -GGRPVAYMSKTLSERAQAKSVYERELMAV 962
PVA+ S+ LS ++++EL+AV
Sbjct: 1423GAAHPVAFHSQRLSPAEYNYPIHDKELLAV 1512
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial (14%)
Length = 1708
Score = 182 bits (463), Expect = 7e-46
Identities = 134/443 (30%), Positives = 217/443 (48%), Gaps = 23/443 (5%)
Frame = +2
Query: 1081 RLLYKNRIVLPKTSGK-IPI------ILQEFHDSAVGGHAGIFRTYKRISALFFWEGMKL 1133
RL ++ RI +P + + P+ ++QE HDS GH G T + +S FFW G
Sbjct: 101 RLTFRGRIWVPGSDDEESPLNELRTKLVQESHDSTAAGHPGRNGTLEIVSRKFFWPGQSQ 280
Query: 1134 DIQTYVQKCEICQRNKYETLNPAGYLQPLPIPSQVWSDISMDFIGGLPKTMGKDT--ILV 1191
++ +V+ C++C G+L+PLP+P+++ SD+SMDFI LP T G+ + + V
Sbjct: 281 TVRRFVRNCDVCGGIHIWRQAKRGFLKPLPVPNRLHSDLSMDFITSLPPTRGRGSQYLWV 460
Query: 1192 VVDRFTKYAHFLALSHPYNAKEVAELFIKEIVKLHGFPTSIVSDRDRVFLSSFWSELFKL 1251
+VDR +K + A+ A+ F+ + HG P SIVSDR ++ FW E +L
Sbjct: 461 IVDRLSKSVTLEEMD-TMEAEACAQRFLSCHYRFHGMPQSIVSDRGSNWVGRFWREFCRL 637
Query: 1252 AGTKLKFSSAYHPQTDGQTEVVNRCVETYLRCVTGAKPKQWPKWLSWAEFWYNTNYHSAI 1311
G S++YHPQTDG TE N+ ++ LR W L + ++S+I
Sbjct: 638 TGVTQLLSTSYHPQTDGGTERWNQEIQAVLRAYVCWSQDNWGDLLPTVQLALRNRHNSSI 817
Query: 1312 KTTPFQALYGR--EPPVIIKGTDSLASVNEVEKMTAERNL--FLDTLKENLEKAQNRMKQ 1367
TPF +G +P ++ T + S E + + ++ + AQ R +
Sbjct: 818 GATPFFVEHGYHVDPIPTVEDTGGVVSEGEAAAQLLVKRMKDVTGFIQAEIVAAQQRSEA 997
Query: 1368 QANKHRRDI-QLKVGDMVYLKIQPYKLKSLARRKNQKLSPRFYGPYPVIEKINAVAYKLQ 1426
ANK R + +VGD V+L + YK + R ++KL + Y V + +L
Sbjct: 998 SANKRRCPADRYQVGDKVWLNVSNYK----SPRPSKKLD-WLHHKYEVTRFVTPHVVELN 1162
Query: 1427 LPEGSQVHPVFHVSLLKKAPN------EGVNSQPLPVYMTEEWELKVEPESIIDSR---A 1477
+P V+P FHV LL++A + E V+ QP P+ + ++ E++ E E I+ +R
Sbjct: 1163 VP--GTVYPKFHVDLLRRAASDPLPGQEVVDPQPPPI-VDDDGEVEWEVEEILAARWHQV 1333
Query: 1478 GDKGTVEVLVKWKNMPEFENSWE 1500
G + LVKWK + +WE
Sbjct: 1334 GRGRRRQALVKWKGF--VDATWE 1396
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 162 bits (409), Expect = 1e-39
Identities = 101/243 (41%), Positives = 136/243 (55%), Gaps = 10/243 (4%)
Frame = +2
Query: 603 LPPKRATDHAILLQEGAPIPNIRPYRYPFYQKNEIEKLV-----KEMLAAGIIRHSTSPF 657
+PP+ D I L +PN+ P P Y+ N ++ V K++L G I+ S P
Sbjct: 17 VPPEWKIDFGIDL-----LPNMNPI*IPSYRINPLKLKVLKLQLKDLLEKGFIQPSIYP* 181
Query: 658 SSPAILVKKKDGGWRFCVDYRALNKVTIPDKFPIPIIDELLDEIGTAEIFSKLDLKSGYH 717
+ +KKKDG R +DY LN V I K+P+P+IDEL D + ++ F K+DL+ G H
Sbjct: 182 GVVVLFLKKKDGFLRMSIDYPQLNNVNIKIKYPLPLIDELFDNLQGSKWFFKIDLRLG*H 361
Query: 718 QIRMREEDIPKTAFRTHEGHYEYLVLPFGLTNAPSTFQALMNQVLRPYLRKFVLVFFYDI 777
Q R+ ED+PKTAFR GHYE LV+ FG TN P F LMN+V + YL V+VF DI
Sbjct: 362 QHRVIGEDVPKTAFRIRYGHYEILVMSFG*TNPPMAFMELMNRVFQDYLDSLVIVFSNDI 541
Query: 778 LIYSNNVDLHKEHLREVLQVLRDNHLVANQKKCSFGQS-ELIYLG----HVISKEGVAAD 832
LIYS N + H+ HLR L+VL+D L C L+ +G HVIS EG+ D
Sbjct: 542 LIYSKNENEHENHLRLALKVLKDIGL------CQISYV*ILVEVGFFSLHVISGEGLKVD 703
Query: 833 PSK 835
+
Sbjct: 704 SKR 712
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 121 bits (304), Expect = 2e-27
Identities = 72/203 (35%), Positives = 109/203 (53%), Gaps = 1/203 (0%)
Frame = +2
Query: 1242 SSFWSELFKLAGTKLKFSSAYHPQTDGQTEVVNRCVETYLRCVTGAKPKQWPKWLSWAEF 1301
S+FW +LFKL GT L SSAYHP +DGQ+E +N+ E YLRC+ P +W K WAE+
Sbjct: 32 SNFWKQLFKLHGTILTMSSAYHP*SDGQSEALNKGXEMYLRCLMFTDPLKWSKAFPWAEY 211
Query: 1302 WYNTNYHSAIKTTPFQALYGREPPVIIKGTDSLASVNEVEKMTAERNLFLDTLKENLEKA 1361
WYNT+Y+ + TPF+ALYGR+ ++I+ S +++ A+R + L L+
Sbjct: 212 WYNTSYNISAAMTPFKALYGRDLSMLIRSKGSSKDTADLQSQLAQR----EELLSQLQSI 379
Query: 1362 QNRM-KQQANKHRRDIQLKVGDMVYLKIQPYKLKSLARRKNQKLSPRFYGPYPVIEKINA 1420
R+ K + K R+ + V + L + R + + ++ +
Sbjct: 380 STRLNKL*SIKLIRNAAILSSSWVSTSL*NCNLINSLR*HCGNIKSSVHP--TLVHY*QS 553
Query: 1421 VAYKLQLPEGSQVHPVFHVSLLK 1443
AYKL LP ++V P+FHVS LK
Sbjct: 554 AAYKLSLPSTAKVPPIFHVSQLK 622
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 92.4 bits (228), Expect(2) = 3e-27
Identities = 47/136 (34%), Positives = 80/136 (58%)
Frame = +2
Query: 721 MREEDIPKTAFRTHEGHYEYLVLPFGLTNAPSTFQALMNQVLRPYLRKFVLVFFYDILIY 780
M +D+ KTAF T G Y Y V+PFGL NA ST+Q L+N++ L + V+ D+L+
Sbjct: 2 MHPDDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVK 181
Query: 781 SNNVDLHKEHLREVLQVLRDNHLVANQKKCSFGQSELIYLGHVISKEGVAADPSKIKDML 840
S H HL+E + L + + N KC+FG + +LG++++++G+ +P +I +L
Sbjct: 182 SLRATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQGIEVNPKQITAIL 361
Query: 841 NWPLPKDVKGLRGFLG 856
+ P PK+ + ++ G
Sbjct: 362 DLPSPKNSREVQRLTG 409
Score = 49.3 bits (116), Expect(2) = 3e-27
Identities = 32/111 (28%), Positives = 54/111 (47%), Gaps = 5/111 (4%)
Frame = +3
Query: 863 RFVRNYSKLAQPLNQLLKKNN-FSWSAGATQAFDKLKEIMTTVPVLAVPDFQKTFVLETD 921
RF+ + P +LL N F W +AF++LK+ +TT PVL+ P+ T L
Sbjct: 429 RFISRSTDKCLPFYKLLCGNKRFVWDEKCEEAFEQLKQYLTTPPVLSKPEAGDTLSLYIA 608
Query: 922 ASGKGLGAVLMQGGR----PVAYMSKTLSERAQAKSVYERELMAVVLAVQK 968
S + +VL++ R P+ Y SK +++ E+ AV+ + +K
Sbjct: 609 ISSTAVSSVLIREDRGEQKPIFYTSKRMTDPETRYPTLEKMAFAVITSARK 761
>BG586326 similar to PIR|G84493|G8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 736
Score = 103 bits (258), Expect = 4e-22
Identities = 75/248 (30%), Positives = 116/248 (46%), Gaps = 29/248 (11%)
Frame = +2
Query: 902 TTVPVLAVPDFQKTFVLETDASGKGLGAVLMQGGRPVAYMSKTLSERAQAKSVYERELMA 961
T+ P+L +P+ T+V+ TDAS GLG VL Q + +AY S+ L + ++ E+ A
Sbjct: 8 TSAPILVLPELI-TYVVYTDASITGLGCVLTQHEKVIAYASRQLRKHEGNYPTHDLEMAA 184
Query: 962 VVLAVQKWRHYLLGCKFIVHTDQKSLRFLAEQRLMGEEQQKWVSKLMGFDFEIKYKPGIE 1021
VV A++ WR YL G K +HTD KSL+++ Q + Q++W+ + +D +I Y PG
Sbjct: 185 VVFALKIWRSYLYGAKVQIHTDHKSLKYIFTQPELNLRQRRWMEFVADYDLDITYYPGKA 364
Query: 1022 NKAADALSRK-------------------LQFSAIS------SVQCEDWEDLETEIL--- 1053
N ADALSR+ L+ + ++ ++ + DL T I
Sbjct: 365 NLVADALSRRRVDVSAEREADDLDGMVRALRLNVLTKATESLGLEAVNQADLFTRIRLAQ 544
Query: 1054 -ADDKYQKIIQEITTQGPVPAGYHMRRGRLLYKNRIVLPKTSGKIPIILQEFHDSAVGGH 1112
D+ QK+ Q T+ + G +L RI +P I+ E H S H
Sbjct: 545 GQDENLQKVAQNDRTEYQT-----AKDGTILVNGRISVPNDRSLKEEIMSEAHKSRFSVH 709
Query: 1113 AGIFRTYK 1120
G R K
Sbjct: 710 PGAPRCIK 733
>BF649369
Length = 631
Score = 87.8 bits (216), Expect = 3e-17
Identities = 44/147 (29%), Positives = 79/147 (52%)
Frame = +3
Query: 88 EHIHHRSHLRAVTGRRVDIPMFNGNDAYGWITKVERFFRLSRVEETEKVEMVMIAMEDRA 147
E++ + + G++V +P+F G+D WIT+ E +F + + +V++ ++ME
Sbjct: 78 EYVESSQNESRLAGKKVKLPLFEGDDPVAWITRAEIYFDVQNTPDDMRVKLSRLSMEGPT 257
Query: 148 LSWFQWWEEQAPVRAWEPFKQALIRRFQPELVQNPFGPLLSAKQKGSVMEYREHFELLAA 207
+ WF E + E K+ALI R+ ++NPF L + +Q GSV E+ E FELL++
Sbjct: 258 IHWFNLLMETEDDLSREKLKKALIARYDGRRLENPFEELSTLRQIGSVEEFVEAFELLSS 437
Query: 208 PMRNADREVLKGVFLNGLHEEIKAEMK 234
+ E G F++GL I+ ++
Sbjct: 438 QVGRLPEEQYLGYFMSGLKAHIRRRVR 518
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 77.8 bits (190), Expect = 3e-14
Identities = 53/174 (30%), Positives = 84/174 (47%), Gaps = 4/174 (2%)
Frame = +2
Query: 1096 KIPIILQEFHDSAVGGHAGIFRTYKRIS-ALFFWEGMKLDIQTYVQKCEICQRNKY---E 1151
+IP IL H S GH + +T +I A F+W M D +++ KC+ CQR
Sbjct: 92 EIPGILFHCHGSNYAGHFAVSKTVSKIQQAGFWWPTMFKDAHSFISKCDPCQRQGNIS*R 271
Query: 1152 TLNPAGYLQPLPIPSQVWSDISMDFIGGLPKTMGKDTILVVVDRFTKYAHFLALSHPYNA 1211
P ++ + + VW +DF+G P + ILV VD +K+ +A S +A
Sbjct: 272 NEMPQNFILEVEV-FDVWG---IDFMGPFPSSYNNKYILVAVDYVSKWVEAIA-SPTNDA 436
Query: 1212 KEVAELFIKEIVKLHGFPTSIVSDRDRVFLSSFWSELFKLAGTKLKFSSAYHPQ 1265
V ++F I G P ++SD F++ + +L K G + K ++AYHPQ
Sbjct: 437 TVVVKMFKSVIFPRFGVPRVVISDGGSHFINKVFEKLLKKNGVRHKVATAYHPQ 598
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 60.8 bits (146), Expect(2) = 4e-12
Identities = 29/63 (46%), Positives = 40/63 (63%)
Frame = -1
Query: 653 STSPFSSPAILVKKKDGGWRFCVDYRALNKVTIPDKFPIPIIDELLDEIGTAEIFSKLDL 712
S SP + + V+KKDG +R C+DYR NKVT +K+P+P ID L D+I F +DL
Sbjct: 256 SISP*GAALLFVRKKDGYFRMCIDYRQFNKVTTKNKYPLPRIDNLFDKIQEDCYF*NIDL 77
Query: 713 KSG 715
+ G
Sbjct: 76 RLG 68
Score = 30.0 bits (66), Expect(2) = 4e-12
Identities = 12/23 (52%), Positives = 15/23 (65%)
Frame = -3
Query: 715 GYHQIRMREEDIPKTAFRTHEGH 737
GYHQ R+ E +IPKT T G+
Sbjct: 71 GYHQFRVNEVNIPKTTLLTWRGY 3
>TC92009
Length = 974
Score = 59.7 bits (143), Expect = 9e-09
Identities = 27/82 (32%), Positives = 44/82 (52%), Gaps = 1/82 (1%)
Frame = +2
Query: 94 SHLRAVTGRRVDIPMFNGNDAYGWITKVERFFRLSRVEETEKVEMVMIAMED-RALSWFQ 152
S + + + + F G D GWIT+ E FF + + +K++ ++MED +AL WF
Sbjct: 653 SEMDGIWTLKYKLSQFTGTDPAGWITRAEMFFADNEIHSCDKLQWAFMSMEDEKALLWFY 832
Query: 153 WWEEQAPVRAWEPFKQALIRRF 174
W ++ P W+ F A+IR F
Sbjct: 833 SWCQENPDADWKSFSMAMIREF 898
>BG644699 similar to PIR|T07863|T078 probable polyprotein - pineapple
retrotransposon dea1 (fragment), partial (5%)
Length = 231
Score = 59.3 bits (142), Expect = 1e-08
Identities = 32/69 (46%), Positives = 43/69 (61%), Gaps = 1/69 (1%)
Frame = +2
Query: 1382 DMVYLKIQPYKLKSLARRKNQKLSPRFYGPYPVIEKINAVAYKLQLPEG-SQVHPVFHVS 1440
+ V LK+ P + K KLS R+ GP+ VI++I VAY+L LP G S VHPVFHVS
Sbjct: 2 EQVLLKVLPTERGDCRFGKRGKLSLRYIGPFEVIKRIGEVAYELALPPGLSGVHPVFHVS 181
Query: 1441 LLKKAPNEG 1449
+ K+ +G
Sbjct: 182 MFKRYHGDG 208
>BF636649 similar to GP|21628724|emb OSJNBa0033H08.7 {Oryza sativa}, partial
(4%)
Length = 653
Score = 56.2 bits (134), Expect = 1e-07
Identities = 52/184 (28%), Positives = 79/184 (42%), Gaps = 15/184 (8%)
Frame = +1
Query: 1266 TDGQTEVVNRCVETYLRCVTGAKPKQWPKWLSWAEFWYNTNYHSAIKTTPFQALYGREPP 1325
+D Q ++N +ET+L T + +L+WAE YNTN+H TPF+ +Y
Sbjct: 7 SDEQAGLLNHTLETHLLYFTSEQQGV*NFFLTWAECLYNTNFHRTAGCTPFKVVY----- 171
Query: 1326 VIIKGTDSLASVNEVEKMTAERNLFLDTLKENLEK---------------AQNRMKQQAN 1370
V ++K R+L E L K A +R +
Sbjct: 172 ----------VVAHLQKFVVARDLIY--RNEGLHKSST*TSFGRGTRAYEALSRPAYETC 315
Query: 1371 KHRRDIQLKVGDMVYLKIQPYKLKSLARRKNQKLSPRFYGPYPVIEKINAVAYKLQLPEG 1430
H R + +VY + + Y+ + L K YGPY VI++I +VA+KL LPE
Sbjct: 316 *HPR----RPLSIVYTRDRTYEWQVL-----PKYVA*CYGPYQVIKQIGSVAFKL*LPEQ 468
Query: 1431 SQVH 1434
Q+H
Sbjct: 469 HQIH 480
>BF003873 similar to GP|14715222|em putative polyprotein {Cicer arietinum},
partial (82%)
Length = 559
Score = 53.1 bits (126), Expect = 8e-07
Identities = 37/117 (31%), Positives = 59/117 (49%), Gaps = 5/117 (4%)
Frame = +2
Query: 1400 KNQKLSPRFYGPYPVIEKINAVAYKLQLPEG-SQVHPVFHVSLLKK-APNEGVNSQPLPV 1457
K++KL+ RF GPY + E++ VAY++ LP +H VFHVS L+K P+ Q V
Sbjct: 20 KSKKLTVRFIGPYQISERVGTVAYRVGLPPHLLNLHDVFHVSQLRKYVPDPSHVIQSDDV 199
Query: 1458 YMTEEWELKVEPESIIDSRA---GDKGTVEVLVKWKNMPEFENSWEDYTELLEQFPE 1511
+ + ++ P I D + K V V W +WE ++++E +PE
Sbjct: 200 QVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVVWDRANGESLTWELESKMVESYPE 370
>TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulchellus},
partial (7%)
Length = 2304
Score = 51.6 bits (122), Expect = 2e-06
Identities = 43/177 (24%), Positives = 75/177 (42%), Gaps = 16/177 (9%)
Frame = +2
Query: 16 ERSITEMKEEAHEQFEELRRLF------MSRERRRTRGRSNTPRHRRVSRDHNSVSLARS 69
ERS+ EM E+ Q ++L+ + + E+RR G ++ S
Sbjct: 431 ERSLQEM-EDMRRQIQQLQEIINAQQALLEAEQRRFEG-----------------DVSYS 556
Query: 70 DDGSRTGSRAVTRSREPHEHIHHRSHLRAVTGRRVDIPMFNGN----DAYGWITKVERFF 125
D S S + R + ++ +VDIP F GN D W+ +ER F
Sbjct: 557 DSSSSRSSHSQRRQLQMNDI-------------KVDIPDFEGNLQLDDFLDWLQTIERVF 697
Query: 126 RLSRVEETEKVEMVMIAMEDRALSWFQ------WWEEQAPVRAWEPFKQALIRRFQP 176
V E +KV++V ++ AL W++ E ++ ++ W+ +Q L R++ P
Sbjct: 698 EYKEVPEEQKVKIVAAKLKKHALIWWENLKRRRKREGKSKIKTWDKMRQKLTRKYLP 868
>BG647713 homologue to GP|15042313|gb| 232R {Chilo iridescent virus}, partial
(1%)
Length = 726
Score = 48.1 bits (113), Expect = 3e-05
Identities = 44/176 (25%), Positives = 77/176 (43%), Gaps = 12/176 (6%)
Frame = +2
Query: 20 TEMKEEAHE-QFEELRRLFMSRERRRTRGRSNTPRHRRVSRDHNSVSLARSDDGSRTGSR 78
T+ + HE + EE+RR + ++ RR DDGS + S
Sbjct: 134 TQNQRPLHEIEMEEMRRQIQLLQETVNAQQALLEAQRR----------RNDDDGSGSDSS 283
Query: 79 AVTRSREPHEHIHHRSHLRAVTGRRVDIPMFNG----NDAYGWITKVERFFRLSRVEETE 134
+ SR HR R ++ +VDIP F G ++ W+ +ER F+ V E +
Sbjct: 284 SSRSSRS------HRRQTR-MSKIKVDIPDF*GKLQPDEFVDWLQTIERVFKYKEVAEEQ 442
Query: 135 KVEMVMIAMEDRALSWF------QWWEEQAPVRAWEPFKQALIRRF-QPELVQNPF 183
KV++V ++ A W+ + E ++ ++ W+ +Q L R++ P Q+ F
Sbjct: 443 KVKIVAAKLKKHASIWWKNLKRKRNCEGKSKIKTWDKMRQKLTRKYLHPHYYQDNF 610
>BG580885
Length = 609
Score = 43.1 bits (100), Expect = 8e-04
Identities = 36/133 (27%), Positives = 61/133 (45%), Gaps = 10/133 (7%)
Frame = +3
Query: 106 IPMFNGNDAYGWITKVERFFRLSRVEETEKVEMVM----IAMEDRALSWFQWWEEQAPVR 161
+P+F G IT + RF ++ R VEM + +E+ + W+ E +
Sbjct: 45 LPIFRGTPNESPITHLSRFNKVCRANNASSVEMQKKIFPVTLEEESALWYDLNIEPYYIS 224
Query: 162 -AWEPFKQALIRRF-QPELVQNPFGPLLSAKQ--KGSVMEY--REHFELLAAPMRNADRE 215
+W+ K + ++ + + E V+ L+ Q K V Y R + L P + +
Sbjct: 225 LSWDEIKLSFLQAYYEIEPVEELRSELMGIHQGEKERVRSYFLRLQWILKRWPEHGLEDD 404
Query: 216 VLKGVFLNGLHEE 228
V+KGVF+NGL EE
Sbjct: 405 VIKGVFVNGLREE 443
>CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product {Drosophila
melanogaster}, partial (20%)
Length = 192
Score = 42.0 bits (97), Expect = 0.002
Identities = 23/58 (39%), Positives = 34/58 (57%), Gaps = 5/58 (8%)
Frame = +1
Query: 926 GLGAVLMQGGR-----PVAYMSKTLSERAQAKSVYERELMAVVLAVQKWRHYLLGCKF 978
G+GA L Q P+AY S+ L+ + +V ERE +A + A++ +RHYL G KF
Sbjct: 13 GIGAGLSQKDEENHEHPIAYASRLLTAAERNYTVVERECLAAIWAIRNFRHYLHGPKF 186
>TC81337 similar to GP|15723293|gb|AAL06332.1 U2 auxiliary factor small
subunit {Arabidopsis thaliana}, partial (81%)
Length = 1190
Score = 38.9 bits (89), Expect = 0.016
Identities = 27/68 (39%), Positives = 35/68 (50%), Gaps = 11/68 (16%)
Frame = +2
Query: 32 ELRRLFMSRERRRTRGRSNTPRHRRVS----------RDHNSVSLARSD-DGSRTGSRAV 80
ELRR +S +R R+R RSN+PR RR S RD++S SD D R G R
Sbjct: 617 ELRRKLVSSQRSRSRSRSNSPRRRRRSRSRERERPRDRDYDSRGRRSSDRDRVRDGDRDR 796
Query: 81 TRSREPHE 88
R R+ +
Sbjct: 797 ERKRDSRD 820
>TC93746 weakly similar to GP|22830935|dbj|BAC15800. hypothetical
protein~similar to gag-pol polyprotein {Oryza sativa
(japonica cultivar-group)}, partial (4%)
Length = 1019
Score = 38.5 bits (88), Expect = 0.021
Identities = 18/42 (42%), Positives = 28/42 (65%), Gaps = 1/42 (2%)
Frame = +2
Query: 1163 PIPSQVWSDISMDFIGGLPKTMG-KDTILVVVDRFTKYAHFL 1203
P+P W D+++DF GL T KD+ +VV D+F++ AHF+
Sbjct: 473 PVPKPPWEDVTIDFSLGLL*TQQLKDSKMVVGDKFSRMAHFI 598
>BG586308 weakly similar to PIR|F84528|F8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 686
Score = 36.2 bits (82), Expect = 0.10
Identities = 22/75 (29%), Positives = 37/75 (49%)
Frame = -2
Query: 1226 HGFPTSIVSDRDRVFLSSFWSELFKLAGTKLKFSSAYHPQTDGQTEVVNRCVETYLRCVT 1285
HG P IV+D F+S+ + E + +L +S +PQ++GQ E N+ + +
Sbjct: 685 HGLPYEIVTDNGSHFISNKFREFCERWRIRLNTASPRYPQSNGQAEASNKII------ID 524
Query: 1286 GAKPKQWPKWLSWAE 1300
G K + K WA+
Sbjct: 523 GLKKRLDLKKGCWAD 479
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 46,733,132
Number of Sequences: 36976
Number of extensions: 669477
Number of successful extensions: 4021
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 3761
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3922
length of query: 1555
length of database: 9,014,727
effective HSP length: 109
effective length of query: 1446
effective length of database: 4,984,343
effective search space: 7207359978
effective search space used: 7207359978
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0045.8


