

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
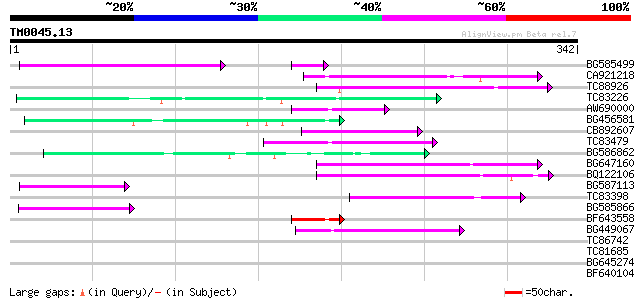
Query= TM0045.13
(342 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG585499 81 4e-17
CA921218 72 4e-13
TC88926 similar to GP|7290507|gb|AAF45960.1| peb gene product {D... 60 2e-09
TC83226 weakly similar to PIR|G86419|G86419 probable reverse tra... 58 6e-09
AW690000 57 1e-08
BG456581 55 4e-08
CB892607 similar to GP|8927657|gb| EST gb|N38213 comes from this... 53 1e-07
TC83479 similar to GP|23476992|emb|CAD48949. hypothetical protei... 53 2e-07
BG586862 51 6e-07
BG647160 homologue to GP|15004435|gb| maturase {Ionopsis satyrio... 50 2e-06
BQ122106 similar to GP|9366656|emb|C probable similar to ring-h2... 49 2e-06
BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At... 49 4e-06
TC83398 similar to PIR|T05150|T05150 hypothetical protein F18E5.... 46 2e-05
BG585866 45 4e-05
BF643558 45 5e-05
BG449067 42 3e-04
TC86742 similar to PIR|T01541|T01541 hypothetical protein A_IG00... 40 0.001
TC81685 40 0.002
BG645274 39 0.004
BF640104 35 0.042
>BG585499
Length = 792
Score = 80.9 bits (198), Expect(2) = 4e-17
Identities = 40/125 (32%), Positives = 58/125 (46%), Gaps = 1/125 (0%)
Frame = +3
Query: 7 WHHIWKWDGPFKVANFLWRVLNNGIWVNYRRWRAGMALDSLCPLCNEESETIIHLLRDCA 66
W +W W GP + F+W V + I NYRR R G + + CP C ET++H+L DC
Sbjct: 225 WKMLWGWRGPHRTQTFMWLVAHGCILTNYRRSRWGTRVLATCPCCGNADETVLHVLCDCR 404
Query: 67 CVRPLWQQLVPGHLFQAFIASDLC-PWLLGQLRSSLLAPIGGGVVPDFGLLLWNIWTSRS 125
+W +LVP F + D C W+ L P F W++WT R+
Sbjct: 405 PASQVWIRLVPSDWITNFFSFDDCRDWVFKNLSKRSNGVSKFKWQPTFMTTCWHMWTWRN 584
Query: 126 CFVFQ 130
+F+
Sbjct: 585 KAIFE 599
Score = 24.3 bits (51), Expect(2) = 4e-17
Identities = 8/22 (36%), Positives = 10/22 (45%)
Frame = +2
Query: 171 VQVGWSPPPPAWVKCNTDGSVR 192
V + W P W K N DG +
Sbjct: 713 VYIAWKRPLDGWAKLNCDGHAK 778
>CA921218
Length = 707
Score = 71.6 bits (174), Expect = 4e-13
Identities = 51/149 (34%), Positives = 67/149 (44%), Gaps = 5/149 (3%)
Frame = -3
Query: 178 PPPAWVKCNTDGSVRGTSNLAACGGVCRDSYGRWLFGFCRNLGTSNVLWAELWGIFSVIQ 237
P P W+ CNT+GS N +ACGG R+S +L F N G N L A+L G I+
Sbjct: 594 PEPLWINCNTNGS--ANINTSACGGTFRNSNAYFLLCFAENTGNGNALHAKLSGAMRAIE 421
Query: 238 LAWDRGVPRLVVESDSSVAVKLINEGSDVLHPYGGMISQIRQWKD-----RDWDFQCVHA 292
+A R L +E DSS+ V S V+H + +W + +F H
Sbjct: 420 IAAARNWSHLWLELDSSLVVNAFKSKS-VIH-----WNLRNRWNNCLFLISSMNFFVSHV 259
Query: 293 CREANSVADELANMAHDLDFGIHILDYPP 321
RE N AD LAN LD + PP
Sbjct: 258 FREGNQCADGLANFGLSLDHLTYWNHVPP 172
>TC88926 similar to GP|7290507|gb|AAF45960.1| peb gene product {Drosophila
melanogaster}, partial (1%)
Length = 1073
Score = 59.7 bits (143), Expect = 2e-09
Identities = 45/146 (30%), Positives = 68/146 (45%), Gaps = 4/146 (2%)
Frame = +3
Query: 186 NTDGSVRGTSNL--AACGGVCRDSYGRWLFGFCRNLGTS-NVLWAELWGIFSVIQLAWDR 242
N DGS+ + A CGGV DS G+WL GF + L + V E I + ++
Sbjct: 351 NVDGSLLREREVPSAGCGGVLSDSSGKWLCGFAQKLNPNLKVDETEKEAILRGLLWVKEK 530
Query: 243 GVPRLVVESDSSVAVKLINEGSDVLHPYGGMISQIRQWKDRDWDFQCVHACREANSVADE 302
G +++V+SD+ V +N G P I + + C+H +N+VAD
Sbjct: 531 GKRKILVKSDNEGVVYSVNCGGRSNDPLVCGIRDLLNSPHWEATLTCIHG--RSNAVADR 704
Query: 303 LANMAHDL-DFGIHILDYPPPRVSNL 327
LA+ AH F + DYPP ++L
Sbjct: 705 LAHKAHSFTSFDLCQFDYPPENCTSL 782
>TC83226 weakly similar to PIR|G86419|G86419 probable reverse transcriptase
100033-105622 [imported] - Arabidopsis thaliana, partial
(2%)
Length = 885
Score = 57.8 bits (138), Expect = 6e-09
Identities = 59/271 (21%), Positives = 102/271 (36%), Gaps = 15/271 (5%)
Frame = +3
Query: 5 LSWHHIWKWDGPFKVANFLWRVLNNGIWVNYRRWRAGMALDSLCPLCNEESETIIHLLRD 64
L W IW + LWR+LN+ + V + G+ LCP C+ ++ETI HL
Sbjct: 114 LIWKKIWSLHTIPRHKVLLWRILNDSLPVRSSLRKRGIQCYPLCPRCHSKTETITHLFMS 293
Query: 65 CACVRPLWQQLVPGHLFQAFIASDLC--------PWLLGQLRSSLLAPIGGGVVPDFGLL 116
C + +W S+LC P + L ++L + +
Sbjct: 294 CPLSKRVW------------FGSNLCINFDNLPNPNFIN*LYEAIL*K-DECITI*IAAI 434
Query: 117 LWNIWTSRSCFVFQGVPFDLHKVLTSGLRQQLDYVHCRNMMQNPAL------PRRASYHT 170
++N+W +R+ V + ++ DY N P++ PR
Sbjct: 435 IYNLWHARNLSVLEDQTILEMDIIQRASNCISDYKQA-NTQAPPSMARTGYDPRSQHRPA 611
Query: 171 VQVGWSPPPPAWVKCNTDGSVRGTSNLAACGGVCRDSYGRWLFGFC-RNLGTSNVLWAEL 229
W P VK NTD +++ G + RD G + G L AE
Sbjct: 612 KNTKWKRPNLGLVKVNTDANLQNHGKW-GLGIIIRDEVGLVMAASTWETDGNDRALEAEA 788
Query: 230 WGIFSVIQLAWDRGVPRLVVESDSSVAVKLI 260
+ + + ++ A D G ++ E D+ +K++
Sbjct: 789 YALLTGMRFAKDCGFXKVXFEGDNEKLMKMV 881
>AW690000
Length = 652
Score = 56.6 bits (135), Expect = 1e-08
Identities = 28/59 (47%), Positives = 35/59 (58%)
Frame = +3
Query: 171 VQVGWSPPPPAWVKCNTDGSVRGTSNLAACGGVCRDSYGRWLFGFCRNLGTSNVLWAEL 229
++V W PP P W+KCNTDGS R S+ +ACGG+ R+ L F N G N AEL
Sbjct: 399 IEVIWRPPIPHWIKCNTDGSSR--SHSSACGGIFRNHDTDLLLCFAENTGECNAFHAEL 569
Score = 30.0 bits (66), Expect = 1.3
Identities = 11/29 (37%), Positives = 19/29 (64%)
Frame = +2
Query: 44 LDSLCPLCNEESETIIHLLRDCACVRPLW 72
L S+C LC +++E+ +HL +C+ LW
Sbjct: 2 LPSMCSLCCKQAESSLHLFFECSYAVNLW 88
>BG456581
Length = 683
Score = 55.1 bits (131), Expect = 4e-08
Identities = 59/208 (28%), Positives = 81/208 (38%), Gaps = 15/208 (7%)
Frame = +2
Query: 10 IWKWDGPFKVANFLWRVLNNGIWVNYRRWRAGMALDSLCPLCNEESETIIHLLRDCACVR 69
IW P + WR+ ++ + + G AL S+C C E+ ET HL C V
Sbjct: 80 IWNSCIPPSHSFICWRLAHDRLPTDDNLSSRGCALVSMCSFCLEQVETSDHLFLRCKFVV 259
Query: 70 PLWQ----QLVPGHLFQAFIASDLCPWLLGQLRSSLLAPIGGGVVPDFGLLLWNIWTSRS 125
LW QL G F +F A LL L + + V ++ +IW +R+
Sbjct: 260 TLWSWLCSQLRVGLDFSSFKA------LLSSLPRHCSSQVRDLYVAAVVHMVHSIWWARN 421
Query: 126 CFVFQGVPFDLHKVLTS-----GLRQQLDYVHC----RNMMQNPALP--RRASYHTVQVG 174
F H V GL + C ++ +P RR+ V V
Sbjct: 422 NVRFSSAKVSAHAVQVRVHALIGLSGAVSTGKCIAADAAILDVFRIPPHRRSMXEMVSVC 601
Query: 175 WSPPPPAWVKCNTDGSVRGTSNLAACGG 202
W PP WVK NTDGS +N A GG
Sbjct: 602 WKPPSAPWVKGNTDGS--XLNNSGAXGG 679
>CB892607 similar to GP|8927657|gb| EST gb|N38213 comes from this gene.
{Arabidopsis thaliana}, partial (1%)
Length = 782
Score = 53.1 bits (126), Expect = 1e-07
Identities = 27/73 (36%), Positives = 39/73 (52%)
Frame = +1
Query: 177 PPPPAWVKCNTDGSVRGTSNLAACGGVCRDSYGRWLFGFCRNLGTSNVLWAELWGIFSVI 236
PP V D V AACGG+ +D G ++F + LG+ +VL AELWGI +
Sbjct: 559 PPNSCEVALKCDYVVLNCDLNAACGGLIQDDQGHFVFHYANKLGSCSVLQAELWGI*HGL 738
Query: 237 QLAWDRGVPRLVV 249
+ W+RG ++ V
Sbjct: 739 SIDWNRGYSKIRV 777
>TC83479 similar to GP|23476992|emb|CAD48949. hypothetical protein {Plasmodium
falciparum 3D7}, partial (0%)
Length = 1222
Score = 52.8 bits (125), Expect = 2e-07
Identities = 33/105 (31%), Positives = 45/105 (42%)
Frame = +2
Query: 154 RNMMQNPALPRRASYHTVQVGWSPPPPAWVKCNTDGSVRGTSNLAACGGVCRDSYGRWLF 213
+N+ Q + S + W P W K NTDGSV A GG+ RD G +
Sbjct: 914 KNLNQIANILNPVSRSIIWCEWKKPEIGWTKLNTDGSVN--KETAGFGGLLRDYRGEPIC 1087
Query: 214 GFCRNLGTSNVLWAELWGIFSVIQLAWDRGVPRLVVESDSSVAVK 258
F + ELW I+ + L+ G+ + VESDS VK
Sbjct: 1088 AFVSKAPQGDTFLVELWAIWRGLVLSLGLGIKSIWVESDSMSVVK 1222
>BG586862
Length = 804
Score = 51.2 bits (121), Expect = 6e-07
Identities = 59/238 (24%), Positives = 95/238 (39%), Gaps = 5/238 (2%)
Frame = -1
Query: 21 NFLWRVLNNGIWVNYRRWRAGMALDSLCPLCNEESETIIHLLRDCACVRPLWQQLVPGHL 80
+FLWR+L+N + V + G+ LCP C + ET+ HL +C + W G
Sbjct: 621 SFLWRLLHNALPVKDELHKRGIRCSLLCPRCESKIETVQHLFLNCEVTQKEWFGSQLGIN 442
Query: 81 FQAFIASDLCPWLLGQLRSSLLAPIGGGVVPDFGLLLWNIWTSRSCFVFQG--VPFDLHK 138
F + W+ ++ + + LL++IW +R+ VF+ VP D+
Sbjct: 441 FHSSGVLHFHDWI-----TNFILKNDEETIIALTALLYSIWHARNQKVFENIDVPGDV-- 283
Query: 139 VLTSGLRQQLDYVHCRNMMQ--NPALPRRASYHTVQVGWSPPPPAWVKCNTDGSVRGTSN 196
+++ +H M Q + LP A P++ S+ G
Sbjct: 282 ----VIQRASSSLHSFKMAQVSDSVLPSNAI------------PSY-------SLWGIGV 172
Query: 197 LAA-CGGVCRDSYGRWLFGFCRNLGTSNVLWAELWGIFSVIQLAWDRGVPRLVVESDS 253
+A C G+ S G WL G AE WGI+ + A D G + ESD+
Sbjct: 171 VARNCEGLAMAS-GTWL-----RHGIPCATTAEAWGIYQAMVFAGDCGFSKFEFESDN 16
>BG647160 homologue to GP|15004435|gb| maturase {Ionopsis satyrioides},
partial (2%)
Length = 780
Score = 49.7 bits (117), Expect = 2e-06
Identities = 38/137 (27%), Positives = 59/137 (42%), Gaps = 1/137 (0%)
Frame = +1
Query: 186 NTDGSVRGTSNLAACGGVCRDSYGRWLFGFCRNLGT-SNVLWAELWGIFSVIQLAWDRGV 244
N + S G GGV R+ ++ GF + N+++AEL + ++LA +
Sbjct: 334 NVERSCIGVPIYT*FGGVIRNYLSTYITGFSGFISIYQNIMFAELTTLHQSLKLAISLNI 513
Query: 245 PRLVVESDSSVAVKLINEGSDVLHPYGGMISQIRQWKDRDWDFQCVHACREANSVADELA 304
+V SDS + V LI E H Y +I I+ + +F H RE N AD +
Sbjct: 514 EEMVCYSDSLLTVNLIKEDISQHHVYAVLIHNIK-YIMSSRNFTLHHTLREGNQCADFMV 690
Query: 305 NMAHDLDFGIHILDYPP 321
+ D + I PP
Sbjct: 691 KLRTSTDVDLTIHSSPP 741
>BQ122106 similar to GP|9366656|emb|C probable similar to ring-h2 finger
protein rha1a. {Trypanosoma brucei}, partial (18%)
Length = 693
Score = 49.3 bits (116), Expect = 2e-06
Identities = 46/146 (31%), Positives = 62/146 (41%), Gaps = 3/146 (2%)
Frame = -2
Query: 186 NTDGSVRGTSNLAACGGVCRDSYGRWLFGFCRNL-GTSNVLWAELWGIFSVIQLAWDRGV 244
N DGS G G+ R+ G + GF N+ TS++L AEL IF +++ D G+
Sbjct: 536 NVDGSCLGNP*PTGFNGLIRNIAGLFNSGFPGNITNTSDILLAELHAIFQGLRMISDMGI 357
Query: 245 PRLVVESDSSVAVKLINEGSDVLHPYGGMISQIRQWKDRDWDFQCVHACREANSVAD--E 302
V DS V LIN S H Y +I I+ H E N AD E
Sbjct: 356 SDFVCYFDSLHYVSLINGPSMKFHVYATLIQDIKDLVITS-KASVFHTLCEGNYCADFLE 180
Query: 303 LANMAHDLDFGIHILDYPPPRVSNLL 328
+ A D IH+ PP + L+
Sbjct: 179 MLGAASDSVLTIHV--SPPDGMIQLI 108
>BG587113 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (10%)
Length = 767
Score = 48.5 bits (114), Expect = 4e-06
Identities = 21/66 (31%), Positives = 34/66 (50%)
Frame = -3
Query: 7 WHHIWKWDGPFKVANFLWRVLNNGIWVNYRRWRAGMALDSLCPLCNEESETIIHLLRDCA 66
+ +WK++ KV +FLWR ++N + ++ D C C ESET+ H+L C
Sbjct: 297 YQRVWKYNTSPKVRHFLWRCISNSLPTAANMRSRHISKDGSCSRCGMESETVNHILFQCP 118
Query: 67 CVRPLW 72
R +W
Sbjct: 117 YARLIW 100
>TC83398 similar to PIR|T05150|T05150 hypothetical protein F18E5.40 -
Arabidopsis thaliana, partial (6%)
Length = 766
Score = 46.2 bits (108), Expect = 2e-05
Identities = 36/107 (33%), Positives = 51/107 (47%), Gaps = 1/107 (0%)
Frame = +2
Query: 206 DSYGRWLFGFCRNLGTSN-VLWAELWGIFSVIQLAWDRGVPRLVVESDSSVAVKLINEGS 264
+ +G + GF ++ SN +L+AEL I +QLA + +V SDS V LIN S
Sbjct: 80 EHFGFFNSGFSGHIDHSNDILFAELHAILMGLQLAQTLNIVDVVCYSDSLHYVNLINGPS 259
Query: 265 DVLHPYGGMISQIRQWKDRDWDFQCVHACREANSVADELANMAHDLD 311
V H Y +I I+ +H RE N AD LA + +D
Sbjct: 260 VVYHAYATLIQDIKDL----IRLSKLHTLREGNRCADFLAKLGASVD 388
>BG585866
Length = 828
Score = 45.1 bits (105), Expect = 4e-05
Identities = 20/70 (28%), Positives = 31/70 (43%)
Frame = +3
Query: 6 SWHHIWKWDGPFKVANFLWRVLNNGIWVNYRRWRAGMALDSLCPLCNEESETIIHLLRDC 65
SW IW+ P K FLW +N + M ++C C E E+ H +RDC
Sbjct: 450 SWSWIWRLKIPEKYKFFLWLACHNAVPTLSLLNHRNMVNSAICSRCGEHEESFFHCVRDC 629
Query: 66 ACVRPLWQQL 75
+ +W ++
Sbjct: 630 RFSKIIWHKI 659
>BF643558
Length = 645
Score = 44.7 bits (104), Expect = 5e-05
Identities = 17/32 (53%), Positives = 24/32 (74%)
Frame = +2
Query: 171 VQVGWSPPPPAWVKCNTDGSVRGTSNLAACGG 202
++V W+PPP W+KCNTDGS +N ++CGG
Sbjct: 545 IEVLWNPPPLHWIKCNTDGS--SNTNTSSCGG 634
>BG449067
Length = 578
Score = 42.4 bits (98), Expect = 3e-04
Identities = 29/103 (28%), Positives = 47/103 (45%), Gaps = 1/103 (0%)
Frame = -2
Query: 173 VGWSPPPPAWVKCNTDGSVRGTSNLAACGGVCRDSYG-RWLFGFCRNLGTSNVLWAELWG 231
V W P +K N+D ++ T +L G V R+ G G G + AE WG
Sbjct: 313 VKWKKPEKDIIKLNSDANLSST-DLWGIGVVARNDEGFAMASGTWFRFGFPSATTAEAWG 137
Query: 232 IFSVIQLAWDRGVPRLVVESDSSVAVKLINEGSDVLHPYGGMI 274
I+ + A + G ++ ESD+ ++++N +V Y G I
Sbjct: 136 IYQAMIFAGEYGFSKVQFESDNERVIQMLNGTEEVNRLYLGSI 8
>TC86742 similar to PIR|T01541|T01541 hypothetical protein A_IG005I10.16 -
Arabidopsis thaliana, partial (19%)
Length = 2073
Score = 40.4 bits (93), Expect = 0.001
Identities = 41/139 (29%), Positives = 60/139 (42%), Gaps = 7/139 (5%)
Frame = -1
Query: 203 VCRDSYGRWLFGFCRNLGTSNVLWAELWGIFSVIQLAWDRGVPRLVVESDSSVAVKLINE 262
V RDS +L N+G + L AE I+ A + G+ + +E+DS +K++N
Sbjct: 468 VIRDSQFGFLGALSCNIGHATPLEAEFCACMIAIEKAMELGLNNICLETDS---LKVVN- 301
Query: 263 GSDVLHPYGGMISQIR-QWKD-----RDWDFQCVHACREANSVADELANMAHDLD-FGIH 315
H G+ Q+R +W + CVH RE N VAD LA L F +
Sbjct: 300 ---AFHKIVGIPWQMRVRWHNCIRFCHSIACVCVHIPREGNLVADALARHGQGLSLFFLQ 130
Query: 316 ILDYPPPRVSNLLLFDVMG 334
PP + + L D G
Sbjct: 129 WWPAPPSFIQSFLAQDRYG 73
>TC81685
Length = 1706
Score = 39.7 bits (91), Expect = 0.002
Identities = 20/49 (40%), Positives = 26/49 (52%)
Frame = -2
Query: 171 VQVGWSPPPPAWVKCNTDGSVRGTSNLAACGGVCRDSYGRWLFGFCRNL 219
+ V W+ P W K N+DG +G S A CGG+ G WL GF + L
Sbjct: 184 ITVSWTLPQSDWDKINSDGMCQG-SLRAGCGGLI*GDGGEWLCGFSKFL 41
>BG645274
Length = 641
Score = 38.5 bits (88), Expect = 0.004
Identities = 15/29 (51%), Positives = 19/29 (64%)
Frame = -3
Query: 178 PPPAWVKCNTDGSVRGTSNLAACGGVCRD 206
P +WVKCN DGS++G A C G+ RD
Sbjct: 369 PILSWVKCNIDGSIKGCFRPATCSGIFRD 283
>BF640104
Length = 344
Score = 35.0 bits (79), Expect = 0.042
Identities = 24/105 (22%), Positives = 46/105 (42%), Gaps = 1/105 (0%)
Frame = -1
Query: 173 VGWSPPPPAWVKCNTDGSVRGTSNLAACGGVCRDSYGRWLF-GFCRNLGTSNVLWAELWG 231
V W P +K N D ++ + ++ G + R+ G + G G + AE WG
Sbjct: 344 VKWIKPHQGVIKINCDANLT-SEDVWGIGVITRNDNGIVMASGTWNRPGFMCPITAEAWG 168
Query: 232 IFSVIQLAWDRGVPRLVVESDSSVAVKLINEGSDVLHPYGGMISQ 276
++ A D+G ++ E+D+ + +++ + Y G I Q
Sbjct: 167 VYQAALFALDQGFQNVLFENDNEKLISMLSREEEGHRSYLGSIIQ 33
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.325 0.141 0.497
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,164,009
Number of Sequences: 36976
Number of extensions: 301116
Number of successful extensions: 1978
Number of sequences better than 10.0: 64
Number of HSP's better than 10.0 without gapping: 1939
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1965
length of query: 342
length of database: 9,014,727
effective HSP length: 97
effective length of query: 245
effective length of database: 5,428,055
effective search space: 1329873475
effective search space used: 1329873475
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0045.13


