

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
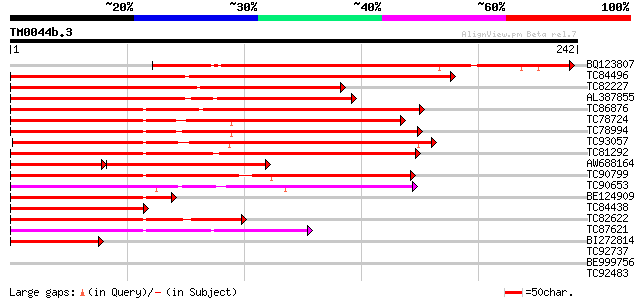
Query= TM0044b.3
(242 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BQ123807 similar to GP|13810202|em MADS2 protein {Cucumis sativu... 194 2e-50
TC84496 similar to GP|1483232|emb|CAA67969.1 MADS5 protein {Betu... 191 3e-49
TC82227 similar to GP|1483232|emb|CAA67969.1 MADS5 protein {Betu... 172 1e-43
AL387855 similar to GP|1483230|emb MADS4 protein {Betula pendula... 156 8e-39
TC86876 similar to GP|16973296|emb|CAC80857. C-type MADS box pro... 154 2e-38
TC78724 similar to GP|4322475|gb|AAD16052.1| putative MADS box t... 149 7e-37
TC78994 similar to SP|O64645|SOC1_ARATH SUPPRESSOR OF CONSTANS O... 147 4e-36
TC93057 homologue to GP|23194453|gb|AAN15183.1 MADS box protein ... 143 5e-35
TC81292 homologue to PIR|T09700|T09700 MADS-box protein - alfalf... 142 9e-35
AW688164 similar to GP|3114588|gb|A MADS box protein {Eucalyptus... 74 2e-29
TC90799 similar to GP|7672991|gb|AAF66690.1| MADS-box transcript... 115 1e-26
TC90653 similar to GP|10835358|gb|AAC13695.2 PTD protein {Populu... 109 1e-24
BE124909 similar to GP|4322475|gb| putative MADS box transcripti... 108 2e-24
TC84438 homologue to GP|1483232|emb|CAA67969.1 MADS5 protein {Be... 100 4e-22
TC82622 similar to GP|21739238|emb|CAD40993. OSJNBa0072F16.17 {O... 91 3e-19
TC87621 similar to SP|Q9FVC1|SVP_ARATH SHORT VEGETATIVE PHASE pr... 91 4e-19
BI272814 PIR|S71757|S71 MADS box protein DEFH200 - garden snapdr... 82 2e-16
TC92737 weakly similar to GP|15022157|gb|AAK77938.1 MADS box pro... 37 0.005
BE999756 weakly similar to GP|9758603|dbj| gene_id:MDF20.13~unkn... 35 0.034
TC92483 similar to GP|21592537|gb|AAM64486.1 unknown {Arabidopsi... 32 0.29
>BQ123807 similar to GP|13810202|em MADS2 protein {Cucumis sativus}, partial
(88%)
Length = 676
Score = 194 bits (494), Expect = 2e-50
Identities = 110/192 (57%), Positives = 131/192 (67%), Gaps = 12/192 (6%)
Frame = +3
Query: 62 SMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGEDLGPL 121
SMLKTL RYQKC+YGA E N +E LE SS +EYLKLKAR+E+LQR+QRNL+GEDLGPL
Sbjct: 6 SMLKTLVRYQKCSYGAVEVNKPAKE-LE-SSYREYLKLKARFESLQRTQRNLLGEDLGPL 179
Query: 122 NSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRLEGYQLNSL 181
+K+LE LERQLDSSLKQ+RST+TQFMLDQL+DLQ KEHML EANRSL +LE +NS
Sbjct: 180 GTKDLEQLERQLDSSLKQVRSTKTQFMLDQLADLQNKEHMLVEANRSLSMKLEEININSR 359
Query: 182 Q-----LNPGVEDMGYGRHPAQTHGDAFYHPIECEPTLQIG----YQPDPVS---VVTAG 229
G + M YG A H +F+ P+EC PTLQIG Y P S T
Sbjct: 360 NQYRQTWEAGDQSMAYGNQNA--HSQSFFQPLECNPTLQIGTDYRYSPPVASDQLTATTQ 533
Query: 230 PSMNNNYMAGWL 241
N ++ GW+
Sbjct: 534 AQQVNGFIPGWM 569
>TC84496 similar to GP|1483232|emb|CAA67969.1 MADS5 protein {Betula
pendula}, partial (74%)
Length = 681
Score = 191 bits (484), Expect = 3e-49
Identities = 101/190 (53%), Positives = 137/190 (71%)
Frame = +2
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
MGRGRV+LK+IENKINRQVTF+KRR+GLLKKA+E+SVLCDAEVALI+FSN+GKLYE+ S
Sbjct: 26 MGRGRVQLKKIENKINRQVTFSKRRSGLLKKAHEISVLCDAEVALIVFSNKGKLYEYSSD 205
Query: 61 SSMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGEDLGP 120
M K LERY++ +Y A +V+ + + E+ +LK R E +Q++QRN MGE+L
Sbjct: 206 PCMEKILERYERYSY-AERQHVANDQPQNENWIIEHARLKTRLEVIQKNQRNFMGEELDG 382
Query: 121 LNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRLEGYQLNS 180
L+ KEL+ LE QLDS+LKQIRS + Q M + +S+L +K+ L E N+ L +++ +
Sbjct: 383 LSMKELQHLEHQLDSALKQIRSRKNQLMYESISELSKKDKALQEKNKLLTTKMKEKEKAL 562
Query: 181 LQLNPGVEDM 190
QL EDM
Sbjct: 563 AQLEQQNEDM 592
>TC82227 similar to GP|1483232|emb|CAA67969.1 MADS5 protein {Betula
pendula}, partial (68%)
Length = 681
Score = 172 bits (435), Expect = 1e-43
Identities = 92/143 (64%), Positives = 113/143 (78%)
Frame = +1
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
MGRGRV+LKRIENKINRQVTF+KRR+GLLKKA E+SVLCDAEVALIIFS +GKL+E+ S
Sbjct: 130 MGRGRVQLKRIENKINRQVTFSKRRSGLLKKAQEISVLCDAEVALIIFSTKGKLFEYSSD 309
Query: 61 SSMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGEDLGP 120
M K LERY++C+Y + V++ ++ + E+ KLKAR E LQR+QRN MGEDL
Sbjct: 310 PCMEKILERYERCSYMERQL-VTSEQSPNENWVLEHAKLKARMEVLQRNQRNFMGEDLDG 486
Query: 121 LNSKELESLERQLDSSLKQIRST 143
L KEL+SLE+QLDS+LKQI T
Sbjct: 487 LGLKELQSLEQQLDSALKQITIT 555
>AL387855 similar to GP|1483230|emb MADS4 protein {Betula pendula}, partial
(51%)
Length = 493
Score = 156 bits (394), Expect = 8e-39
Identities = 84/148 (56%), Positives = 113/148 (75%)
Frame = +2
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
MGRGRV+LKRIENK ++QVTF KRR GLLKKA E+SVLCDA+VALI+FS +GKL+E+ S+
Sbjct: 53 MGRGRVQLKRIENKTSQQVTFFKRRTGLLKKANEISVLCDAQVALIMFSTKGKLFEYSSA 232
Query: 61 SSMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGEDLGP 120
SM LERY++ N+ E +T E + + EY+KL A+ + L+R+ RN +G DL P
Sbjct: 233 PSMEDILERYERQNH--TELTGATNET-QGNWSFEYMKLTAKVQVLERNLRNFVGNDLDP 403
Query: 121 LNSKELESLERQLDSSLKQIRSTRTQFM 148
L+ KEL+SLE+QLD+SLK+IR+ + Q M
Sbjct: 404 LSVKELQSLEQQLDTSLKRIRTRKNQVM 487
>TC86876 similar to GP|16973296|emb|CAC80857. C-type MADS box protein {Malus
x domestica}, partial (85%)
Length = 2050
Score = 154 bits (390), Expect = 2e-38
Identities = 83/177 (46%), Positives = 121/177 (67%)
Frame = +2
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
MGRG++E+KRIEN NRQVTF KRRNGLLKKAYELSVLCDAEVAL++FS RG+LYE+ ++
Sbjct: 89 MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALVVFSTRGRLYEY-AN 265
Query: 61 SSMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGEDLGP 120
+S+ T+ERY+K + A S EA QQE KL+ + +Q R+++GE LG
Sbjct: 266 NSVRATIERYKKACAASTNAE-SVSEANTQFYQQESSKLRRQIRDIQNLNRHILGEALGS 442
Query: 121 LNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRLEGYQ 177
L+ KEL++LE +L+ L ++RS + + + + +Q++E L N LR ++ ++
Sbjct: 443 LSLKELKNLEGRLEKGLSRVRSRKHETLFADVEFMQKREIELQNHNNYLRAKIAEHE 613
>TC78724 similar to GP|4322475|gb|AAD16052.1| putative MADS box
transcription factor ETL {Eucalyptus globulus subsp.
globulus}, partial (71%)
Length = 1089
Score = 149 bits (377), Expect = 7e-37
Identities = 85/172 (49%), Positives = 120/172 (69%), Gaps = 3/172 (1%)
Frame = +1
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
M RG+ ++KRIEN+ NRQVTF+KRRNGLLKKA+ELSVLCDAEVALI+FS GKLYEF SS
Sbjct: 226 MVRGKTQMKRIENESNRQVTFSKRRNGLLKKAFELSVLCDAEVALIVFSTTGKLYEF-SS 402
Query: 61 SSMLKTLERYQKCNYGAPEANVSTREALELSSQ---QEYLKLKARYEALQRSQRNLMGED 117
SS+ KT+ERYQ G + V + + ++ ++Q + + + E L+ S+R L+GE+
Sbjct: 403 SSISKTVERYQ----GKVKELVLSTKGIQENTQHLKECDIDTTKKLEHLELSKRKLLGEE 570
Query: 118 LGPLNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSL 169
LG EL+ +E QL+ SL +IR+ + Q + +Q+ L+ KE +L E N+ L
Sbjct: 571 LGSCAFDELQQIENQLERSLSKIRARKNQLLKEQIEKLKDKERLLLEENKRL 726
>TC78994 similar to SP|O64645|SOC1_ARATH SUPPRESSOR OF CONSTANS
OVEREXPRESSION 1 protein (Agamous-like MADS box protein
AGL20)., partial (83%)
Length = 946
Score = 147 bits (371), Expect = 4e-36
Identities = 82/178 (46%), Positives = 122/178 (68%), Gaps = 2/178 (1%)
Frame = +3
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
M RG+ ++KRIEN +RQVTF+KRRNGLLKKA+ELSVLCDAEVALI+FS RG+LYEF +S
Sbjct: 54 MVRGKTQMKRIENATSRQVTFSKRRNGLLKKAFELSVLCDAEVALIVFSPRGRLYEF-AS 230
Query: 61 SSMLKTLERYQKCNYGAPEANVSTREALELSSQ--QEYLKLKARYEALQRSQRNLMGEDL 118
SS+L+T+ERY+ ++ +T E++E + Q +E + + + L+ S+R L+GE L
Sbjct: 231 SSILETIERYR--SHTRINNTPTTSESVENTQQLKEEAENMMKKIDLLETSKRKLLGEGL 404
Query: 119 GPLNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRLEGY 176
G + EL+ +E+QL+ S+ +IR +T+ +Q+ L+ KE L N L ++ Y
Sbjct: 405 GSCSIDELQKIEQQLEKSINKIRVKKTKVFREQIDQLKEKEKALVAENVRLSEKYGNY 578
>TC93057 homologue to GP|23194453|gb|AAN15183.1 MADS box protein GHMADS-2
{Gossypium hirsutum}, partial (95%)
Length = 858
Score = 143 bits (361), Expect = 5e-35
Identities = 86/196 (43%), Positives = 124/196 (62%), Gaps = 15/196 (7%)
Frame = +3
Query: 2 GRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSSS 61
G R+ K IEN NRQVTF KRRNGLLKKAYELSVLCDAEVALI+FS+RG+LYE+ S++
Sbjct: 33 GGERLR*KGIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEY-SNN 209
Query: 62 SMLKTLERYQKCNYGAPEANVSTREALELSS---QQEYLKLKARYEALQRSQRNLMGEDL 118
++ T++RY+K A + ST E+++ QQE KL+ + + LQ S R+LMG+ L
Sbjct: 210 NIRSTIDRYKK----ACSDHSSTTTTTEINAQYYQQESAKLRQQIQMLQNSNRHLMGDAL 377
Query: 119 GPLNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRL----- 173
L KEL+ LE +L+ + +IRS + + +L ++ Q++E L N LR ++
Sbjct: 378 STLTVKELKQLENRLERGITRIRSKKHEMLLAEIEYFQKREIELENENLCLRTKINDVER 557
Query: 174 -------EGYQLNSLQ 182
G +LN++Q
Sbjct: 558 LPQVNMVSGQELNAIQ 605
>TC81292 homologue to PIR|T09700|T09700 MADS-box protein - alfalfa
(fragment), partial (91%)
Length = 800
Score = 142 bits (359), Expect = 9e-35
Identities = 76/175 (43%), Positives = 115/175 (65%)
Frame = +2
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
MGRG+++++RI+N +RQVTF+KRRNGLLKKA EL++LCDAEV ++IFS+ KLY+F +S
Sbjct: 71 MGRGKIQIRRIDNSTSRQVTFSKRRNGLLKKAKELAILCDAEVGVMIFSSTAKLYDF-AS 247
Query: 61 SSMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGEDLGP 120
+SM ++RY K + ST E Q+E L+ + LQ S R +MGE+L
Sbjct: 248 TSMRSVIDRYNKTKEEHNQLGSSTSEI--KFWQREAAMLRQQLHNLQESHRQIMGEELSG 421
Query: 121 LNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRLEG 175
L KEL+ LE QL+ SL+ +R + Q +D++ +L RK ++ + N L +++ G
Sbjct: 422 LTVKELQGLENQLEISLRGVRMKKEQLFMDEIQELNRKGDIIHQENVELYRKVYG 586
>AW688164 similar to GP|3114588|gb|A MADS box protein {Eucalyptus grandis},
partial (31%)
Length = 549
Score = 73.6 bits (179), Expect(2) = 2e-29
Identities = 35/70 (50%), Positives = 50/70 (71%)
Frame = +2
Query: 42 EVALIIFSNRGKLYEFCSSSSMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKA 101
EVALIIFS+RGKL+++ S++ + K +ERY++C Y P+A S + Q+YLKLKA
Sbjct: 131 EVALIIFSSRGKLFQYSSTTDINKIIERYRQCRYSKPQAGNSLGHNESQNLYQDYLKLKA 310
Query: 102 RYEALQRSQR 111
+YE+L R QR
Sbjct: 311 KYESLDRKQR 340
Score = 72.8 bits (177), Expect(2) = 2e-29
Identities = 35/41 (85%), Positives = 39/41 (94%)
Frame = +1
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDA 41
MGRGRV L+RIENKINRQVTF+KRR+GLLKKA+EL VLCDA
Sbjct: 7 MGRGRVVLERIENKINRQVTFSKRRSGLLKKAFELCVLCDA 129
>TC90799 similar to GP|7672991|gb|AAF66690.1| MADS-box transcription factor
{Canavalia lineata}, partial (76%)
Length = 653
Score = 115 bits (289), Expect = 1e-26
Identities = 67/175 (38%), Positives = 108/175 (61%), Gaps = 2/175 (1%)
Frame = +2
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
M R ++++K+I+N RQVTF+KRR G+ KKA ELS+LCDAEV L+IFS GKLYE+ +S
Sbjct: 71 MARQKIKIKKIDNATARQVTFSKRRRGIFKKAEELSILCDAEVGLVIFSTTGKLYEY-AS 247
Query: 61 SSMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRSQ--RNLMGEDL 118
S+M + RY + ++ + + + +E + E K E R+Q R + ED
Sbjct: 248 SNMKDIITRYGQQSHHITKLDKPLQVQVEKNMPAELNK-----EVADRTQQLRGMKSEDF 412
Query: 119 GPLNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRL 173
LN + L+ LE+ L+S LK++ + + +L+++ L+ KE ML N+ L+Q++
Sbjct: 413 EGLNLEGLQQLEKSLESXLKRVIEMKEKKILNEIKALRMKEIMLEXENKHLKQKM 577
>TC90653 similar to GP|10835358|gb|AAC13695.2 PTD protein {Populus
balsamifera subsp. trichocarpa}, partial (59%)
Length = 845
Score = 109 bits (272), Expect = 1e-24
Identities = 69/178 (38%), Positives = 107/178 (59%), Gaps = 4/178 (2%)
Frame = +2
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
MGRG++E+K IEN NRQVT++KRRNG+ KKA+ELSVLCDA+V+LI+FS K++E+ +
Sbjct: 44 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 223
Query: 61 S-SMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGE--- 116
S K +++YQK G + S E + + KLK L+R R+ +GE
Sbjct: 224 GLSTKKIIDQYQK-TLGDIDLWRSHYEKM----LENLKKLKDINHKLRRQIRHRIGEGGM 388
Query: 117 DLGPLNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRLE 174
+L L+ ++L SLE ++SS+ +IR + + + ++K L + N +L LE
Sbjct: 389 ELDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLELE 562
>BE124909 similar to GP|4322475|gb| putative MADS box transcription factor
ETL {Eucalyptus globulus subsp. globulus}, partial (60%)
Length = 608
Score = 108 bits (269), Expect = 2e-24
Identities = 55/71 (77%), Positives = 64/71 (89%)
Frame = +1
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
M RG+ ++KRIEN+ NRQVTF+KRRNGLLKKA+ELSVLCDAEVALI+FS GKLYEF SS
Sbjct: 148 MVRGKTQMKRIENESNRQVTFSKRRNGLLKKAFELSVLCDAEVALIVFSTTGKLYEF-SS 324
Query: 61 SSMLKTLERYQ 71
SS+ KT+ERYQ
Sbjct: 325 SSISKTVERYQ 357
>TC84438 homologue to GP|1483232|emb|CAA67969.1 MADS5 protein {Betula
pendula}, partial (24%)
Length = 702
Score = 100 bits (250), Expect = 4e-22
Identities = 49/59 (83%), Positives = 56/59 (94%)
Frame = +1
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCS 59
MGRGRV+LKRIENKINRQVTF+KRR+GLLKKA E+SVLCDAEVALIIFS +GKL+E+ S
Sbjct: 223 MGRGRVQLKRIENKINRQVTFSKRRSGLLKKAQEISVLCDAEVALIIFSTKGKLFEYSS 399
>TC82622 similar to GP|21739238|emb|CAD40993. OSJNBa0072F16.17 {Oryza
sativa}, partial (78%)
Length = 847
Score = 91.3 bits (225), Expect = 3e-19
Identities = 48/101 (47%), Positives = 71/101 (69%)
Frame = +3
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
MGRG++ ++RI+N +RQVTF+KRR GL+KKA EL++LCDA+V L+IFS+ GKLYE+ ++
Sbjct: 138 MGRGKIVIRRIDNCTSRQVTFSKRRKGLIKKAKELAILCDAQVGLVIFSSTGKLYEY-AN 314
Query: 61 SSMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKA 101
+SM +ERY C + V+ E+ Q+E + KA
Sbjct: 315 TSMKSVIERYNICK---EDQQVTNPESEVKFWQREAVHFKA 428
>TC87621 similar to SP|Q9FVC1|SVP_ARATH SHORT VEGETATIVE PHASE protein.
[Mouse-ear cress] {Arabidopsis thaliana}, partial (65%)
Length = 1322
Score = 90.9 bits (224), Expect = 4e-19
Identities = 54/129 (41%), Positives = 77/129 (58%)
Frame = +2
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
M R ++++K+IEN RQVTF+KRR GL+KKA ELSVLCDA+VALIIFS+ GKL+E+ S+
Sbjct: 437 MAREKIQIKKIENSTARQVTFSKRRRGLIKKAEELSVLCDADVALIIFSSTGKLFEY-SN 613
Query: 61 SSMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGEDLGP 120
SM + LER+ + + + E L+L +L R + GEDL
Sbjct: 614 LSMREILERHHLHSKNLAKLEEPSLE-LQLVENSNCSRLSKEVAQKSHQLRQMRGEDLQG 790
Query: 121 LNSKELESL 129
S + ++
Sbjct: 791 FESGRVTTI 817
>BI272814 PIR|S71757|S71 MADS box protein DEFH200 - garden snapdragon,
partial (16%)
Length = 320
Score = 82.0 bits (201), Expect = 2e-16
Identities = 40/40 (100%), Positives = 40/40 (100%)
Frame = +1
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCD 40
MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCD
Sbjct: 199 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCD 318
>TC92737 weakly similar to GP|15022157|gb|AAK77938.1 MADS box protein-like
protein NGL9 {Medicago sativa}, partial (33%)
Length = 541
Score = 37.4 bits (85), Expect = 0.005
Identities = 22/77 (28%), Positives = 41/77 (52%)
Frame = +3
Query: 98 KLKARYEALQRSQRNLMGEDLGPLNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQR 157
++K +++Q R+L GED+ LN KEL +LE L++ L +R + ++ +R
Sbjct: 24 RIKKENDSMQIDLRHLKGEDITSLNYKELMALEESLENGLTGVRDKK----MEVHRMFKR 191
Query: 158 KEHMLSEANRSLRQRLE 174
+L + N+ L L+
Sbjct: 192 NGKILEDENKELNFLLQ 242
>BE999756 weakly similar to GP|9758603|dbj| gene_id:MDF20.13~unknown
protein {Arabidopsis thaliana}, partial (15%)
Length = 534
Score = 34.7 bits (78), Expect = 0.034
Identities = 17/48 (35%), Positives = 26/48 (53%)
Frame = +1
Query: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIF 48
M +V+L I N R+ T+ KR L+KK E++ LC + I+F
Sbjct: 13 MAGNKVKLAFITNHTARRATYKKRVQSLMKKLNEITTLCGVKACGIVF 156
>TC92483 similar to GP|21592537|gb|AAM64486.1 unknown {Arabidopsis
thaliana}, partial (8%)
Length = 662
Score = 31.6 bits (70), Expect = 0.29
Identities = 21/77 (27%), Positives = 37/77 (47%)
Frame = +2
Query: 60 SSSMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGEDLG 119
+S+ + T+++ + G+ EA++S L S L+ ALQ +QR + L
Sbjct: 416 NSTKISTIKKSKSFKEGSTEASLSYSSNLITDSPGSIAGLRREQMALQNAQRKMKDCSLW 595
Query: 120 PLNSKELESLERQLDSS 136
+ + ES+ LDSS
Sbjct: 596 KIKVCKFESVSIPLDSS 646
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.132 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,405,500
Number of Sequences: 36976
Number of extensions: 78758
Number of successful extensions: 378
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 365
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 366
length of query: 242
length of database: 9,014,727
effective HSP length: 93
effective length of query: 149
effective length of database: 5,575,959
effective search space: 830817891
effective search space used: 830817891
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0044b.3


