

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
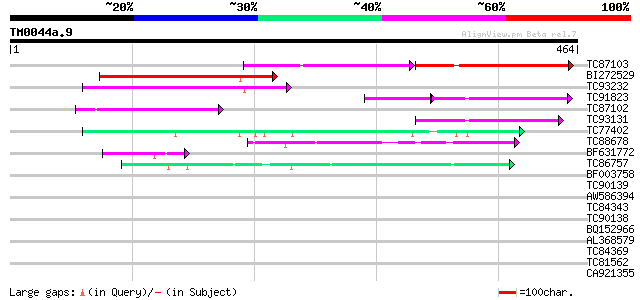
Query= TM0044a.9
(464 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87103 similar to GP|15292735|gb|AAK92736.1 unknown protein {Ar... 90 4e-39
BI272529 weakly similar to GP|6478925|gb| hypothetical protein {... 110 1e-24
TC93232 weakly similar to GP|21739227|emb|CAD40982. OSJNBa0072F1... 94 8e-20
TC91823 weakly similar to GP|19310729|gb|AAL85095.1 unknown prot... 62 2e-18
TC87102 similar to PIR|T02589|T02589 hypothetical protein T16B24... 85 5e-17
TC93131 similar to GP|19310729|gb|AAL85095.1 unknown protein {Ar... 73 3e-13
TC77402 similar to PIR|T50691|T50691 amino acid permease 6 [impo... 60 2e-09
TC88678 similar to PIR|S51171|S51171 amino acid transport protei... 46 3e-05
BF631772 similar to PIR|F86385|F86 hypothetical protein AAG12739... 44 1e-04
TC86757 auxin influx carrier protein [Medicago truncatula] 42 5e-04
BF003758 similar to GP|15216030|em amino acid permease AAP4 {Vic... 39 0.003
TC90139 similar to GP|15290010|dbj|BAB63704. contains ESTs C9952... 39 0.003
AW586394 similar to GP|15216026|em amino acid permease AAP1 {Vic... 39 0.004
TC84343 auxin influx carrier protein [Medicago truncatula] 38 0.009
TC90138 similar to GP|15290010|dbj|BAB63704. contains ESTs C9952... 38 0.009
BQ152966 similar to GP|14194151|gb AT5g40780/K1B16_3 {Arabidopsi... 38 0.009
AL368579 similar to GP|14194151|gb AT5g40780/K1B16_3 {Arabidopsi... 37 0.012
TC84369 similar to GP|19310729|gb|AAL85095.1 unknown protein {Ar... 35 0.047
TC81562 weakly similar to GP|6671946|gb|AAF23206.1| putative ami... 34 0.10
CA921355 weakly similar to GP|452593|dbj| ORF {Lilium longifloru... 34 0.14
>TC87103 similar to GP|15292735|gb|AAK92736.1 unknown protein {Arabidopsis
thaliana}, partial (51%)
Length = 1621
Score = 90.1 bits (222), Expect(2) = 4e-39
Identities = 51/130 (39%), Positives = 80/130 (61%), Gaps = 1/130 (0%)
Frame = +1
Query: 333 IALWATVLTPMTKYALEFAPVAIQLEHKLPSSMSGRTKMILRGI-IGSSLLLVILALALT 391
IA+W TV+ P TKYAL +PVA+ LE +P++ K L I I + L+ L + L+
Sbjct: 943 IAVWTTVVNPFTKYALTISPVAMSLEELIPAN---HAKSYLFSIFIRTGLVFSTLVIGLS 1113
Query: 392 VPYFEHVLSLTGSLVSVAISLIFPCAFYIKICRGQISKPLFVLNISIITFGFLLGVVGTI 451
VP+F V+SL GSL+++ ++LI PC Y++I RG++++ L I+II G VGT
Sbjct: 1114 VPFFGLVMSLIGSLLTMLVTLILPCVCYLRILRGKVTRIQAGLCITIIVVGVACSSVGTY 1293
Query: 452 SSSKLLVEKI 461
S+ +V+ +
Sbjct: 1294 SALAEIVKSL 1323
Score = 89.7 bits (221), Expect(2) = 4e-39
Identities = 52/141 (36%), Positives = 76/141 (53%), Gaps = 1/141 (0%)
Frame = +3
Query: 192 PQIL-TVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSVAATAVFGGVRVNHEIPF 250
PQ L V A L LP++WLRDLS +S++S+ GV+ SV++ + + + V
Sbjct: 519 PQTLFAVVAALAVLPTVWLRDLSVLSYISAGGVIASVLVVLCLLWIGI-EDVGFQRSGTT 695
Query: 251 LRLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFTKVSILSFTTVTALYTIMGYMGAK 310
L L +P G+Y + Y GH VFPN+Y +M P++F V + F T LY MG K
Sbjct: 696 LNLGTLPVAIGLYGYCYSGHAVFPNIYTSMAKPNQFPAVLVACFGVCTLLYAGGAVMGYK 875
Query: 311 MFGPEVNSQVTLSMPSKQIVT 331
G + SQ TL++P + T
Sbjct: 876 NVGEDTLSQFTLNLPQDLVAT 938
>BI272529 weakly similar to GP|6478925|gb| hypothetical protein {Arabidopsis
thaliana}, partial (23%)
Length = 625
Score = 110 bits (274), Expect = 1e-24
Identities = 58/148 (39%), Positives = 93/148 (62%), Gaps = 2/148 (1%)
Frame = +2
Query: 74 SLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCTYTCHLLGKCLKKNPKL 133
S A SV+N +L G+G ++ PYAV++GG+ S LL+ V+C YT LL +CL+ P L
Sbjct: 134 SFARSVINGTNLLCGIGLMTMPYAVKEGGFLSLALLLLFAVICCYTGILLMRCLQSYPGL 313
Query: 134 RSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISLHDNLTTVLHLKQLHLA--KLSA 191
+Y DIG AFG GR AI++ +++F + V + + DNL+ + + +A +LS
Sbjct: 314 DTYPDIGQAAFGYAGRLGIAIILSLELFGASVEFLTLVSDNLSALFPNTSMIVAGTELST 493
Query: 192 PQILTVGAVLVALPSLWLRDLSSISFLS 219
Q+ T+ A L+ LP+ WL++LS +S++S
Sbjct: 494 HQVFTIAAALLVLPTGWLKNLSLLSYIS 577
>TC93232 weakly similar to GP|21739227|emb|CAD40982. OSJNBa0072F16.6 {Oryza
sativa}, partial (56%)
Length = 695
Score = 94.4 bits (233), Expect = 8e-20
Identities = 51/175 (29%), Positives = 96/175 (54%), Gaps = 4/175 (2%)
Frame = +3
Query: 60 DTNVDAEHDAKANS-SLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCTY 118
+ V H + N+ S H+ +N + + G+G LS PYA+ GGW S LL + Y
Sbjct: 120 EEKVIVSHPSNKNTVSFFHTCVNGLNAISGVGILSVPYALASGGWLSLALLFCIAAAAFY 299
Query: 119 TCHLLGKCLKKNPKLRSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISLHDNLTTV 178
+ L+ +C++KN +++Y DIG AFG GR + +I +Y ++++ + + I DNL+ +
Sbjct: 300 SGILMKRCMEKNSNIKTYPDIGELAFGKIGRLIVSISMYTELYLVSIGFLILEGDNLSNL 479
Query: 179 LHLKQLHLAKLS--APQILTVGAVLVALPSLWLRDL-SSISFLSSVGVLMSVVIF 230
+++ + LS A + + ++ LP++WL + S+ + V L+ V++F
Sbjct: 480 FPIEEFQVFGLSIGAKKFFVILVAVIILPTIWLDNFESAFLWYLQVVCLLLVLLF 644
>TC91823 weakly similar to GP|19310729|gb|AAL85095.1 unknown protein
{Arabidopsis thaliana}, partial (20%)
Length = 926
Score = 62.4 bits (150), Expect(2) = 2e-18
Identities = 35/115 (30%), Positives = 65/115 (56%)
Frame = +2
Query: 346 YALEFAPVAIQLEHKLPSSMSGRTKMILRGIIGSSLLLVILALALTVPYFEHVLSLTGSL 405
YAL P+A LE LP S+S L ++ ++L++ + A +P+F V++L GSL
Sbjct: 206 YALMMNPLARSLEELLPDSISRTNWCFL--LLRTALVISTVCAAFLIPFFGLVMALIGSL 379
Query: 406 VSVAISLIFPCAFYIKICRGQISKPLFVLNISIITFGFLLGVVGTISSSKLLVEK 460
+SV ++++ P ++KI + + +L++ I FG + +GT SS +V++
Sbjct: 380 LSVLVAMVLPAFCFLKIVGKRATNKQVILSVVIAAFGIVCASLGTYSSLLKIVKR 544
Score = 47.8 bits (112), Expect(2) = 2e-18
Identities = 21/58 (36%), Positives = 32/58 (54%)
Frame = +3
Query: 291 ILSFTTVTALYTIMGYMGAKMFGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYAL 348
+ SF LY +G G MFG +SQ+TL +P +K++LW + P+TK+ L
Sbjct: 39 LYSFVLPVFLYGSVGAAGFLMFGERTSSQITLDLPRDAFASKVSLWTIAIIPLTKHML 212
>TC87102 similar to PIR|T02589|T02589 hypothetical protein T16B24.23 -
Arabidopsis thaliana (fragment), partial (44%)
Length = 964
Score = 85.1 bits (209), Expect = 5e-17
Identities = 43/121 (35%), Positives = 69/121 (56%)
Frame = +2
Query: 55 SAVARDTNVDAEHDAKANSSLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIALGV 114
S + +++ V E ++ + S +V+N + +L G+G LSTPYA ++GGW +L G+
Sbjct: 557 SLLKKESKVSHEVPSR-HCSFGQAVLNGINVLCGVGILSTPYAAKEGGWLGLSILFIFGI 733
Query: 115 MCTYTCHLLGKCLKKNPKLRSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISLHDN 174
+ YT LL CL P L +Y DIG AFG GR +I++Y++++ + Y DN
Sbjct: 734 LSFYTGLLLRSCLDSEPGLETYPDIGQAAFGTAGRIAISIVLYVELYGCCIEYINLEGDN 913
Query: 175 L 175
L
Sbjct: 914 L 916
>TC93131 similar to GP|19310729|gb|AAL85095.1 unknown protein {Arabidopsis
thaliana}, partial (22%)
Length = 786
Score = 72.8 bits (177), Expect = 3e-13
Identities = 37/121 (30%), Positives = 70/121 (57%)
Frame = +2
Query: 333 IALWATVLTPMTKYALEFAPVAIQLEHKLPSSMSGRTKMILRGIIGSSLLLVILALALTV 392
++LW + P+TKYAL P+A +E LP S+S + ++ ++L++ + A +
Sbjct: 2 VSLWTIAVIPLTKYALMMNPLARSVEELLPHSISSTNWCFI--LLRTALVISTVCAAFVI 175
Query: 393 PYFEHVLSLTGSLVSVAISLIFPCAFYIKICRGQISKPLFVLNISIITFGFLLGVVGTIS 452
P+F V++L GSL+SV ++++ P ++KI + + VL++ I +G + +GT S
Sbjct: 176 PFFGLVMALIGSLLSVLVAIVLPALCFLKIVGKKATSTQVVLSVVIAAWGVVCASLGTYS 355
Query: 453 S 453
S
Sbjct: 356 S 358
>TC77402 similar to PIR|T50691|T50691 amino acid permease 6 [imported] -
Arabidopsis thaliana, partial (90%)
Length = 1902
Score = 60.1 bits (144), Expect = 2e-09
Identities = 92/416 (22%), Positives = 160/416 (38%), Gaps = 54/416 (12%)
Frame = +3
Query: 60 DTNVDAEHDAKANSSLAHSVMNMVGMLIGLGQLSTPYAVEQGGW-ASAILLIALGVMCTY 118
D N D + AK + + +++ +IG G LS +A+ Q GW A +L A + +
Sbjct: 327 DKNFDDDGRAKRTGTWLTASAHIITAVIGSGVLSLAWAIAQMGWVAGPAVLFAFSFITYF 506
Query: 119 TCHLLGKCLKKNPKLR-----SYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISLHD 173
T LL C + + +Y ++ G + L + Y+++ + YTI+
Sbjct: 507 TSTLLADCYRSPDPVHGKRNYTYTEVVRANLGGRKFQLCGLAQYINLVGVTIGYTITASI 686
Query: 174 NLTTVLHLKQLHL------AKLSAPQILTVGA----VLVALPSL----WLRDLSSI-SFL 218
++ V H +S + + A VL +P+ WL ++++ SF
Sbjct: 687 SMVAVQRSNCFHKHGHQDKCYVSNNPFMIIFACIQIVLCQIPNFHELSWLSIVAAVMSFA 866
Query: 219 -SSVGVLMSVVIF------VSVAATAVFGGVRVNHEIPFLRLHN-IPSISGIYIFSYGGH 270
SS+G+ +SV V+ + T V GV V R+ I I+ Y FS
Sbjct: 867 YSSIGLGLSVAKVAGGGNHVTTSLTGVQIGVDVTATEKVWRMFQAIGDIAFAYAFSNVLI 1046
Query: 271 IVFPNLYKAMKDPSKFTKVSILSFTTVTALYTIMGYMGAKMFGPEVNSQVTLSMPSKQ-- 328
+ L + + + S++ T T Y + G +G FG + +
Sbjct: 1047EIQDTLKSSPPENRVMKRASLIGILTTTLFYVLCGTLGYAAFGNDAPGNFLTGFGFYEPF 1226
Query: 329 ----------IVTKIALWATVLTPMTKYALEFAPVAIQLEHKLPSS--MSGRTKMIL--- 373
V I + + P+ F V Q + K P S ++G M +
Sbjct: 1227WLIDFANVCIAVHLIGAYQVFVQPI------FGFVEGQSKQKWPDSKFVNGEHAMNIPLY 1388
Query: 374 --------RGIIGSSLLLVILALALTVPYFEHVLSLTGSLVSVAISLIFPCAFYIK 421
R I S +++ +A+ P+F L L GSL +++ FP YIK
Sbjct: 1389GSYNVNYFRVIWRSCYVIITAIIAMLFPFFNDFLGLIGSLSFYPLTVYFPIEMYIK 1556
>TC88678 similar to PIR|S51171|S51171 amino acid transport protein AAT1 -
Arabidopsis thaliana, partial (72%)
Length = 1520
Score = 46.2 bits (108), Expect = 3e-05
Identities = 50/227 (22%), Positives = 98/227 (43%), Gaps = 4/227 (1%)
Frame = +3
Query: 195 LTVGAVLVALPSLWLRDLSSISFLSSVGVL--MSVVIFVSVAATAVFGGVRVNHEIPFLR 252
+ +GA LVA+ +L + S + + + M+V+IF+ +A N PF
Sbjct: 654 IAIGA-LVAITALAVYSTKGSSIFNYIATMFHMAVIIFIVIAGLIKAKPENFNDFTPF-G 827
Query: 253 LHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFTKVSIL-SFTTVTALYTIMGYMGAKM 311
LH + S S + F+Y G + + K+P + + ++ S T TA+Y ++G
Sbjct: 828 LHGMVSSSAVLFFAYIGFDAVSTMAEETKNPGRDIPIGLVGSMTITTAIYCLLG------ 989
Query: 312 FGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYALEFAPVAIQLEHKLPSSMSGRTKM 371
+ + L K++ T A + +++A + L ++ G T +
Sbjct: 990 ------ATLCLMQNYKELDTD----APFSVAFSAVGMDWAKYIVSL-----GALKGMTTV 1124
Query: 372 ILRGIIGSSLLLVILALA-LTVPYFEHVLSLTGSLVSVAISLIFPCA 417
+L +G + L +A + P+F V TG+ ++ IS++ A
Sbjct: 1125LLVSAVGQARYLTHIARTHMMPPWFALVDERTGTPMNATISMLIATA 1265
>BF631772 similar to PIR|F86385|F86 hypothetical protein AAG12739.1
[imported] - Arabidopsis thaliana, partial (29%)
Length = 523
Score = 44.3 bits (103), Expect = 1e-04
Identities = 26/75 (34%), Positives = 38/75 (50%), Gaps = 4/75 (5%)
Frame = +2
Query: 77 HSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCT----YTCHLLGKCLKKNPK 132
+S + V +IG G LS PYA+ GW IL++ L T + L +C+ +
Sbjct: 125 YSTFHTVTAMIGAGVLSLPYAMAYLGWGPGILMLLLSWCLTLNTMWQMIQLHECV-PGTR 301
Query: 133 LRSYVDIGNHAFGAK 147
Y+D+G HAFG K
Sbjct: 302 FDRYIDLGRHAFGPK 346
>TC86757 auxin influx carrier protein [Medicago truncatula]
Length = 1590
Score = 42.0 bits (97), Expect = 5e-04
Identities = 71/337 (21%), Positives = 134/337 (39%), Gaps = 15/337 (4%)
Frame = +1
Query: 92 LSTPYAVEQGGWASAILL-IALGVMCTYTCHLLGKCLK--KNPKLRSYVDIGNHAF---- 144
L+ PY+ Q G S IL I G+M ++T +++ + K R VD NH
Sbjct: 271 LTLPYSFSQLGMLSGILFQIFYGLMGSWTAYIISVLYVEYRTRKEREKVDFRNHVIQWFE 450
Query: 145 ---GAKGRFLAAILIYMDIFMSLVSYTISLHDNLTTVLHLKQLHLAKLSAPQIL-TVGAV 200
G G+ + ++ + L I L + + ++ HL K + I A
Sbjct: 451 VLDGLLGKHWRNLGLFFNCTFLLFGSVIQLIACASNIYYIND-HLDKRTWTYIFGACCAT 627
Query: 201 LVALPSLWLRDLSSISFLSSVGVLMSVVI--FVSVAATAVFGGVRVNHEIPFLRLHNIPS 258
V +PS + S +G++M+ ++++A+ V H P +
Sbjct: 628 TVFIPSF-----HNYRIWSFLGLVMTTYTAWYMTIASILHGQAEDVKHSGPTKLVLYFTG 792
Query: 259 ISGIYIFSYGGHIVFPNLYKAMKDPSKFTKVSILSFTTVTALYTIMGYMGAKMFGPEVNS 318
+ I ++++GGH V + AM P KF + +++ V L FG + +
Sbjct: 793 ATNI-LYTFGGHAVTVEIMHAMWKPQKFKMIYLIATLYVMTLTLPSAAAVYWAFGDNLLT 969
Query: 319 QVT-LSMPSKQIVTKIALWATVLTPMTKYALEFAPVAIQLEHKLPSSMSGRTKMILRGII 377
LS+ + A+ ++ + P+ E L + ++ R ++
Sbjct: 970 HSNALSLLPRTGFRDTAVILMLIHQFITFGFACTPLYFVWEKFL--GVHETKSLLKRALV 1143
Query: 378 GSSLLLVILALALTVPYFEHVLSLTGS-LVSVAISLI 413
+++ I LA+ P+F + S GS LVS + +I
Sbjct: 1144RLPVVIPIWFLAIIFPFFGPINSTVGSLLVSFTVYII 1254
>BF003758 similar to GP|15216030|em amino acid permease AAP4 {Vicia faba var.
minor}, partial (31%)
Length = 554
Score = 39.3 bits (90), Expect = 0.003
Identities = 33/130 (25%), Positives = 60/130 (45%), Gaps = 19/130 (14%)
Frame = +2
Query: 60 DTNVDAEHDAK-------------ANSSLAHSVMNMVGMLIGLGQLSTPYAVEQGGW-AS 105
D ++D + D+K A ++ AH ++ +IG G LS +A+ Q GW A
Sbjct: 137 DVSIDQQRDSKFFDDDGRVKRTGTAWTASAH----VITAVIGSGVLSLAWAIAQLGWIAG 304
Query: 106 AILLIALGVMCTYTCHLLGKCLKKNPKLR-----SYVDIGNHAFGAKGRFLAAILIYMDI 160
I++I + YT LL +C + + +Y+D+ + G L I+ Y+++
Sbjct: 305 PIVMILFAWVTYYTSILLCECYRTGDPISGKRNYTYMDVVHSNLGGFQVTLCGIVQYLNL 484
Query: 161 FMSLVSYTIS 170
+ YTI+
Sbjct: 485 VGVAIGYTIA 514
>TC90139 similar to GP|15290010|dbj|BAB63704. contains ESTs C99526(E20351)
C73742(E20351)~unknown protein {Oryza sativa (japonica
cultivar-group)}, partial (26%)
Length = 680
Score = 39.3 bits (90), Expect = 0.003
Identities = 28/117 (23%), Positives = 54/117 (45%), Gaps = 6/117 (5%)
Frame = +2
Query: 59 RDTNVDAEHDAKANSSLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCT- 117
RD + A K+ S H ++ ++ LS PYA GW + I + +G M T
Sbjct: 158 RDVDAGALFVLKSKGSWVHCGYHLTTSIVAPPLLSLPYAFTLLGWTAGIFFLVIGAMVTF 337
Query: 118 YTCHLLGKCLKKNPKLRS----YVDIGNHAFGAK-GRFLAAILIYMDIFMSLVSYTI 169
Y+ +LL + L+ +L + + D+ G + GR+ + + + ++V+ T+
Sbjct: 338 YSYNLLSRVLEHQAQLGNRQLRFRDMARDILGPRWGRYFVGPIQFAVCYGAVVACTL 508
>AW586394 similar to GP|15216026|em amino acid permease AAP1 {Vicia faba var.
minor}, partial (42%)
Length = 613
Score = 38.9 bits (89), Expect = 0.004
Identities = 37/162 (22%), Positives = 66/162 (39%), Gaps = 14/162 (8%)
Frame = +1
Query: 274 PNLYKAMKDPSKFTKVSILSFTTVTALYTIMGYMGAKMFGPEVNSQVTLSMPSKQIVTKI 333
P+ K MK +K +S TA Y + G MG FG + + + + I
Sbjct: 145 PSEVKTMKQATK------ISIGVTTAFYMLCGCMGYAAFGDTAPGNLLTGIFNPYWLIDI 306
Query: 334 ALWATVLTPMTKYALEFAPV-------------AIQLEHKLPSSMSGRTKMILRGIIGSS 380
A A V+ + Y + P I E+++P + L +I +
Sbjct: 307 ANAAIVIHLVGAYQVYAQPFFAFVEKIVIKRWPKINKEYRIPIPGFHPYNLNLFRLIWRT 486
Query: 381 LLLVILA-LALTVPYFEHVLSLTGSLVSVAISLIFPCAFYIK 421
+ ++ +A+ +P+F VL L G++ +++ FP YIK
Sbjct: 487 IFVITTTVIAMLIPFFNDVLGLLGAVGFWPLTVYFPVEMYIK 612
>TC84343 auxin influx carrier protein [Medicago truncatula]
Length = 1579
Score = 37.7 bits (86), Expect = 0.009
Identities = 80/416 (19%), Positives = 154/416 (36%), Gaps = 36/416 (8%)
Frame = +1
Query: 67 HDAKANSSLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILL-IALGVMCTYTCHLLGK 125
H A + N V ++ L+ PY+ Q G S IL + G++ ++T +L+
Sbjct: 118 HGGSAYDAWFSCASNQVAQVL----LTLPYSFSQLGMLSGILFQLFYGILGSWTAYLISI 285
Query: 126 CLK--KNPKLRSYVDIGNHAF-------GAKGRFLAAILIYMDIFMSLVSYTISLHDNLT 176
+ K R V+ +H G G+ + + + L I L +
Sbjct: 286 LYVEYRTRKEREKVNFRSHVIQWFEVLDGLLGKHWRNVGLGFNCTFLLFGSVIQLIACAS 465
Query: 177 TVLHLKQLHLAKLSAPQIL-TVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSVAA 235
+ ++ +L K + I A V +PS + S +G++M+ +
Sbjct: 466 NIYYIND-NLDKRTWTYIFGACCATTVFIPSF-----HNYRIWSFLGLVMTTYTAWYLTI 627
Query: 236 TAVFGGV--RVNHEIPFLRLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFTKVSILS 293
AV G V H P + + I ++++GGH V + AM P KF + +L+
Sbjct: 628 AAVLHGQVEGVKHSGPNKIILYFTGATNI-LYTFGGHAVTVEIMHAMWKPQKFKAIYLLA 804
Query: 294 --------FTTVTALYTIMGYMGAKMFGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTK 345
+ TA+Y G M +N ++ K +A+ ++
Sbjct: 805 TLYVLTLTIPSATAVYWAFGDM-------LLNHSNAFALLPKSPFRDMAVILMLIHQFIT 963
Query: 346 YALEFAPVAIQLEHKLPSSMSGRTKMILRGIIGSSLLLVILALALTVPYFEHVLSLTGSL 405
+ P+ E + M + R ++ +++ I LA+ P+F + S GSL
Sbjct: 964 FGFACTPLYFVWEKTV--GMHECKSLCKRALVRLPVVIPIWFLAIIFPFFGPINSTVGSL 1137
Query: 406 VSVAISLIFPCAFYIKICRGQISKP---------------LFVLNISIITFGFLLG 446
+ I P +I + ++ FV+N+ I+ + ++G
Sbjct: 1138LVSFTVYIIPALAHIFTFKSSSARQNAVEQPPKFVGRWVGTFVINVFIVVWVLIVG 1305
>TC90138 similar to GP|15290010|dbj|BAB63704. contains ESTs C99526(E20351)
C73742(E20351)~unknown protein {Oryza sativa (japonica
cultivar-group)}, partial (30%)
Length = 573
Score = 37.7 bits (86), Expect = 0.009
Identities = 22/76 (28%), Positives = 36/76 (46%), Gaps = 1/76 (1%)
Frame = +3
Query: 59 RDTNVDAEHDAKANSSLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCT- 117
RD + A K+ S H ++ ++ LS PYA GW + I + +G M T
Sbjct: 141 RDVDAGALFVLKSKGSWVHCGYHLTTSIVAPPLLSLPYAFTLLGWTAGIFFLVIGAMVTF 320
Query: 118 YTCHLLGKCLKKNPKL 133
Y+ +LL + L+ +L
Sbjct: 321 YSYNLLSRVLEHQAQL 368
>BQ152966 similar to GP|14194151|gb AT5g40780/K1B16_3 {Arabidopsis thaliana},
partial (31%)
Length = 574
Score = 37.7 bits (86), Expect = 0.009
Identities = 25/122 (20%), Positives = 51/122 (41%)
Frame = +2
Query: 339 VLTPMTKYALEFAPVAIQLEHKLPSSMSGRTKMILRGIIGSSLLLVILALALTVPYFEHV 398
V+ + Y + PV +E L M+ +LR I+ + + + +A+T P+F+ +
Sbjct: 29 VIHVIGSYQIYAMPVFDMIETLLVKKMNFEPSTMLRFIVRNVYVAFTMFIAITFPFFDGL 208
Query: 399 LSLTGSLVSVAISLIFPCAFYIKICRGQISKPLFVLNISIITFGFLLGVVGTISSSKLLV 458
L G + PC ++ I + + + N I G L ++ I + ++
Sbjct: 209 LGFFGGFAFAPTTYFLPCIMWLSIYKPRKFGLSWWANWICIVLGVCLMILSPIGGLRTII 388
Query: 459 EK 460
K
Sbjct: 389 IK 394
>AL368579 similar to GP|14194151|gb AT5g40780/K1B16_3 {Arabidopsis thaliana},
partial (29%)
Length = 504
Score = 37.4 bits (85), Expect = 0.012
Identities = 24/79 (30%), Positives = 39/79 (48%), Gaps = 3/79 (3%)
Frame = +1
Query: 72 NSSLAHSVMNMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCT-YTCHLLGKCLKKN 130
N+ +S + V ++G G LS PYA+ Q GW + ++ L T YT + + +
Sbjct: 154 NAKWWYSAFHNVTAMVGAGVLSLPYAMAQLGWGPEVTVLVLSWFITLYTLWQMVEMHEMV 333
Query: 131 PKLR--SYVDIGNHAFGAK 147
P R Y ++ +AFG K
Sbjct: 334 PGKRFDRYHELSQYAFGEK 390
>TC84369 similar to GP|19310729|gb|AAL85095.1 unknown protein {Arabidopsis
thaliana}, partial (8%)
Length = 614
Score = 35.4 bits (80), Expect = 0.047
Identities = 16/46 (34%), Positives = 26/46 (55%), Gaps = 1/46 (2%)
Frame = +3
Query: 74 SLAHSVMNMVGMLIGLGQLSTPYAVEQGGWAS-AILLIALGVMCTY 118
+ ++ N + ++ G+G LSTP V+Q GW S ++LI M Y
Sbjct: 423 TFTQTIFNGINIMAGVGLLSTPDTVKQAGWVSLVVMLIFCSCMLXY 560
>TC81562 weakly similar to GP|6671946|gb|AAF23206.1| putative amino acid
transporter protein {Arabidopsis thaliana}, partial
(36%)
Length = 673
Score = 34.3 bits (77), Expect = 0.10
Identities = 35/165 (21%), Positives = 62/165 (37%), Gaps = 7/165 (4%)
Frame = +1
Query: 112 LGVMCTYTCHLLGKCLKKNPKL--RSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTI 169
L V +T K + P + RSY D+G +G GR+L +I + ++Y +
Sbjct: 172 LSVFILFTVWCRDKLASEEPLVESRSYGDLGYRCYGILGRYLTEFIIVVAQCAGAIAYLV 351
Query: 170 SLHDNLTTVLHLKQLHLAKLSAPQILTVGAVLVALPSLWLRDLSSIS----FLSSVGVLM 225
+ NL +V LA V V + W+ LS+++ F VL
Sbjct: 352 FIGQNLDSVFQKYGPSLAS------YIFMLVPVEIGMSWIGSLSALAPFSIFADVCNVLA 513
Query: 226 SVVIFVSVAATAVFGGVRVNHEIPFL-RLHNIPSISGIYIFSYGG 269
++ A+ G + +P +G+ +F + G
Sbjct: 514 MGIVVKEDVQVALEDGFSFRERTAITSNIGGLPFAAGMAVFCFEG 648
>CA921355 weakly similar to GP|452593|dbj| ORF {Lilium longiflorum}, partial
(22%)
Length = 505
Score = 33.9 bits (76), Expect = 0.14
Identities = 15/72 (20%), Positives = 40/72 (54%)
Frame = -1
Query: 388 LALTVPYFEHVLSLTGSLVSVAISLIFPCAFYIKICRGQISKPLFVLNISIITFGFLLGV 447
+ + +P+ + L G +++ ++L +PC ++K+ + + ++ LN + TFG L V
Sbjct: 397 IGVAIPFLSSLAGLIGG-IALPVTLAYPCFMWLKVKKPKKYSFMWYLNWFLGTFGVALSV 221
Query: 448 VGTISSSKLLVE 459
+ +S ++++
Sbjct: 220 ILVTASIYVIID 185
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.324 0.137 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,605,615
Number of Sequences: 36976
Number of extensions: 213851
Number of successful extensions: 1553
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 1530
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1547
length of query: 464
length of database: 9,014,727
effective HSP length: 100
effective length of query: 364
effective length of database: 5,317,127
effective search space: 1935434228
effective search space used: 1935434228
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0044a.9


