

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
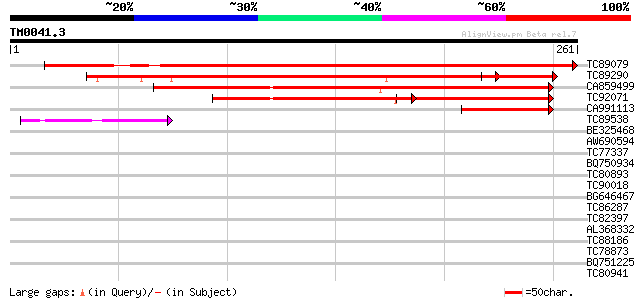
Query= TM0041.3
(261 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC89079 similar to GP|20805123|dbj|BAB92794. hypothetical protei... 358 e-100
TC89290 similar to GP|3738315|gb|AAC63656.1| unknown protein {Ar... 267 2e-81
CA859499 weakly similar to PIR|T41359|T41 hypothetical protein S... 209 1e-54
TC92071 weakly similar to PIR|T41359|T41359 hypothetical protein... 103 9e-23
CA991113 homologue to PIR|G86387|G863 hypothetical protein AAG29... 72 2e-13
TC89538 weakly similar to GP|21689895|gb|AAM67508.1 unknown prot... 42 3e-04
BE325468 similar to GP|10727848|gb Eip75B gene product {Drosophi... 39 0.003
AW690594 similar to GP|23498163|emb hypothetical protein {Plasmo... 38 0.004
TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Atel... 38 0.004
BQ750934 similar to PIR|T19673|T19 hypothetical protein C33B4.3 ... 37 0.008
TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {... 37 0.010
TC90018 weakly similar to PIR|B84607|B84607 hypothetical protein... 36 0.017
BG646467 similar to SP|O64668|PSNH Presenilin homolog. [Mouse-ea... 35 0.022
TC86287 similar to GP|23617108|dbj|BAC20790. contains ESTs AU056... 35 0.029
TC82397 similar to GP|18377877|gb|AAL67124.1 AT3g53630/F4P12_330... 35 0.038
AL368332 weakly similar to GP|21322711|e pherophorin-dz1 protein... 35 0.038
TC88186 similar to GP|15293177|gb|AAK93699.1 unknown protein {Ar... 35 0.038
TC78873 similar to GP|21740530|emb|CAD41509. OSJNBa0029H02.25 {O... 34 0.050
BQ751225 similar to GP|18376350|em probable aldehyde dehydrogena... 34 0.050
TC80941 weakly similar to PIR|T39903|T39903 serine-rich protein ... 34 0.065
>TC89079 similar to GP|20805123|dbj|BAB92794. hypothetical protein {Oryza
sativa (japonica cultivar-group)}, partial (38%)
Length = 753
Score = 358 bits (920), Expect = e-100
Identities = 189/245 (77%), Positives = 200/245 (81%)
Frame = +2
Query: 17 IPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSASSAI 76
I SS SS QLR K + S HM AA SS+TT PS PS SSAI
Sbjct: 38 ISSSSSSAQLRLKPPSSSSLGARVCCHMAHAA-------SSITTPSPS-----PSPSSAI 181
Query: 77 DFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGVDRDKCVKMAIVH 136
DFLSICHRLKTTKRAGW+RK+VQ+PESIADHMYRM LMALIA PGVDRDKCVKMAIVH
Sbjct: 182 DFLSICHRLKTTKRAGWLRKEVQNPESIADHMYRMGLMALIAPHFPGVDRDKCVKMAIVH 361
Query: 137 DIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKEVAELWAEYEANSSPEA 196
DIAEAIVGDITP DG+PKEEK+R EQEALDHMCKVLGGGSRAKE+AELW EYEANSSPEA
Sbjct: 362 DIAEAIVGDITPIDGIPKEEKSRREQEALDHMCKVLGGGSRAKEIAELWTEYEANSSPEA 541
Query: 197 KFVKDLDKVEMILQALEYEHEQGKDLDEFFQSTAGKFQTEIGKAWASEIVSRRKKTNGSS 256
KFVKDLDKVEMILQALEYE EQ KDLDEFF+STAGKFQTEIGK WA+EIVSRR KTN SS
Sbjct: 542 KFVKDLDKVEMILQALEYEEEQEKDLDEFFRSTAGKFQTEIGKTWAAEIVSRRNKTNDSS 721
Query: 257 HPHST 261
H +ST
Sbjct: 722 HSNST 736
>TC89290 similar to GP|3738315|gb|AAC63656.1| unknown protein {Arabidopsis
thaliana}, partial (65%)
Length = 1106
Score = 267 bits (683), Expect(2) = 2e-81
Identities = 134/203 (66%), Positives = 165/203 (81%), Gaps = 12/203 (5%)
Frame = +3
Query: 36 HSPS--RVFHMTTAAGSSSPPSSSVT----TSPPSAASATPSAS----SAIDFLSICHRL 85
H+P+ R M TA +SS PS++VT T+PP ++SA P AS +AI+FL++CH L
Sbjct: 54 HTPTFTRNHLMATAPSASSSPSTAVTGDTNTTPPPSSSAVPDASPSPFAAIEFLTVCHHL 233
Query: 86 KTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGVDRDKCVKMAIVHDIAEAIVGD 145
K TKRAGW+ +DV+DPES+ADHMYRMSLMALIA D+PG+DR+KC+KMAIVHDIAEA++GD
Sbjct: 234 KKTKRAGWVTRDVKDPESVADHMYRMSLMALIAPDVPGLDRNKCIKMAIVHDIAEALIGD 413
Query: 146 ITPTDGVPKEEKNRLEQEALDHMCKVL--GGGSRAKEVAELWAEYEANSSPEAKFVKDLD 203
ITP DGV K +K+ EQ AL++MCK++ G GSR KE+ ELW +YEANSSPEAKFVKDLD
Sbjct: 414 ITPLDGVSKADKSEREQAALEYMCKIIGVGEGSRGKEITELWMDYEANSSPEAKFVKDLD 593
Query: 204 KVEMILQALEYEHEQGKDLDEFF 226
KVEMILQAL+YE EQGKDLDE F
Sbjct: 594 KVEMILQALDYEDEQGKDLDEIF 662
Score = 52.8 bits (125), Expect(2) = 2e-81
Identities = 25/35 (71%), Positives = 29/35 (82%)
Frame = +1
Query: 218 QGKDLDEFFQSTAGKFQTEIGKAWASEIVSRRKKT 252
+G+ +FF STAGKFQTE+GKAWASEIVSRR T
Sbjct: 637 KGRIWMKFFCSTAGKFQTEVGKAWASEIVSRRSNT 741
>CA859499 weakly similar to PIR|T41359|T41 hypothetical protein SPCC4G3.17 -
fission yeast (Schizosaccharomyces pombe), partial (61%)
Length = 575
Score = 209 bits (531), Expect = 1e-54
Identities = 100/185 (54%), Positives = 137/185 (74%), Gaps = 1/185 (0%)
Frame = +3
Query: 67 SATPSASSAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGVDR 126
+ T + S+ ++FL IC +LK TKR GWI D+++PESI+DHM+RMS+++L+ D ++R
Sbjct: 18 NTTTNTSNILEFLHICEKLKKTKRTGWICHDIKNPESISDHMHRMSILSLLIKD-SSLNR 194
Query: 127 DKCVKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMC-KVLGGGSRAKEVAELW 185
DKCVKMA+VHD+AEA+VGDITP +G+ K EKNR E EA+ MC +L +++E+ LW
Sbjct: 195 DKCVKMAVVHDLAEALVGDITPFEGISKAEKNRREFEAMRQMCTNLLENSPQSQEIFALW 374
Query: 186 AEYEANSSPEAKFVKDLDKVEMILQALEYEHEQGKDLDEFFQSTAGKFQTEIGKAWASEI 245
EYE NS+ EAKFVKD+DK EMI+QA EYE + KDL++FF+ST GKF K W E+
Sbjct: 375 QEYEDNSTNEAKFVKDIDKFEMIIQAYEYEQSENKDLEQFFESTKGKFNHPEVKCWVEEL 554
Query: 246 VSRRK 250
S+RK
Sbjct: 555 YSKRK 569
>TC92071 weakly similar to PIR|T41359|T41359 hypothetical protein SPCC4G3.17
- fission yeast (Schizosaccharomyces pombe), partial
(37%)
Length = 752
Score = 103 bits (256), Expect = 9e-23
Identities = 51/105 (48%), Positives = 75/105 (70%), Gaps = 11/105 (10%)
Frame = +3
Query: 94 IRKDVQDPESIADHMYRMSLMALIAADIPGVDRDKCVKMAIVHDIAEAIVGDITPTDGVP 153
I+ ++++PESI+DHMYRMS++AL+ D +D++KCVKMA+VHD+AE IVGDI+P +G+
Sbjct: 18 IQHNIKNPESISDHMYRMSILALLLND-DDLDKNKCVKMAVVHDLAEGIVGDISPAEGIT 194
Query: 154 KEEKNRLEQEALDHMCKVLGGGS-----------RAKEVAELWAE 187
KEEK+R E EA+ H+CK L S R ++A+LW +
Sbjct: 195 KEEKHRREMEAMQHICKKLLENSLQVSRRFFLCGRNMKMAKLWRQ 329
Score = 84.7 bits (208), Expect = 3e-17
Identities = 40/72 (55%), Positives = 50/72 (68%)
Frame = +2
Query: 179 KEVAELWAEYEANSSPEAKFVKDLDKVEMILQALEYEHEQGKDLDEFFQSTAGKFQTEIG 238
KE+ LW EYE + EAKFVKDLDK E+ILQA EYE + KDL EFF++T GKF
Sbjct: 275 KEIFSLWQEYENGETMEAKFVKDLDKFELILQAFEYEKSEKKDLSEFFENTRGKFNHPHV 454
Query: 239 KAWASEIVSRRK 250
K+WA E+ +R+
Sbjct: 455 KSWAEELYLKRE 490
>CA991113 homologue to PIR|G86387|G863 hypothetical protein AAG29220.1
[imported] - Arabidopsis thaliana, partial (17%)
Length = 369
Score = 72.4 bits (176), Expect = 2e-13
Identities = 35/42 (83%), Positives = 38/42 (90%)
Frame = +1
Query: 209 LQALEYEHEQGKDLDEFFQSTAGKFQTEIGKAWASEIVSRRK 250
LQALEYE E GK LDEFF STAGKFQTEIGK+WA+EI+SRRK
Sbjct: 1 LQALEYELEHGKVLDEFFISTAGKFQTEIGKSWAAEIISRRK 126
>TC89538 weakly similar to GP|21689895|gb|AAM67508.1 unknown protein
{Arabidopsis thaliana}, partial (37%)
Length = 990
Score = 41.6 bits (96), Expect = 3e-04
Identities = 29/70 (41%), Positives = 37/70 (52%)
Frame = +3
Query: 6 RVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSA 65
++L S FS + +S SS RF VT P S +TAA ++ P S T SPP
Sbjct: 99 QILRSRFFS--VAASSSSKPFRFSKVTPQPSSR----FSSTAAPATPHPPPSATPSPPPP 260
Query: 66 ASATPSASSA 75
+SATP SSA
Sbjct: 261 SSATPPPSSA 290
>BE325468 similar to GP|10727848|gb Eip75B gene product {Drosophila
melanogaster}, partial (1%)
Length = 434
Score = 38.5 bits (88), Expect = 0.003
Identities = 18/45 (40%), Positives = 26/45 (57%)
Frame = +2
Query: 38 PSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSASSAIDFLSIC 82
PS +FH+T + G+ PP S TS S +S+ S+SS+ S C
Sbjct: 290 PSEIFHLTLSYGNDDPPESLARTSTSSRSSSASSSSSSSSTSSNC 424
>AW690594 similar to GP|23498163|emb hypothetical protein {Plasmodium
falciparum 3D7}, partial (10%)
Length = 633
Score = 37.7 bits (86), Expect = 0.004
Identities = 28/77 (36%), Positives = 44/77 (56%)
Frame = -3
Query: 6 RVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSA 65
RVL+ S+ SSS SS SS+ S + S S S ++++ SSS SSS ++S S+
Sbjct: 271 RVLILSISSSSPSSSSSSSS---SSSSSSSSSSSSSCSSSSSSSSSSSSSSSCSSSSSSS 101
Query: 66 ASATPSASSAIDFLSIC 82
S++ S++S S+C
Sbjct: 100 CSSSSSSTSPSSSSSLC 50
Score = 32.0 bits (71), Expect = 0.25
Identities = 24/82 (29%), Positives = 44/82 (53%)
Frame = -3
Query: 1 MATATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTT 60
+ AT VL+ SI ++ S ++ S++ S S S ++++ SSS SSS ++
Sbjct: 337 LIVATEVLVGR--KESICNNSGSGRVLILSISSSSPSSSSSSSSSSSSSSSSSSSSSCSS 164
Query: 61 SPPSAASATPSASSAIDFLSIC 82
S S++S++ S+S + S C
Sbjct: 163 SSSSSSSSSSSSSCSSSSSSSC 98
Score = 31.6 bits (70), Expect = 0.32
Identities = 25/62 (40%), Positives = 33/62 (52%)
Frame = -3
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASAT 69
SS SSS SS SS+ S + S S S +++ SSS SSS +TSP S++S
Sbjct: 229 SSSSSSSSSSSSSSSSSSSCSSSSSSSSSSSSSSSCSSSSSSSCSSSSSSTSPSSSSSLC 50
Query: 70 PS 71
S
Sbjct: 49 SS 44
>TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Ateline
herpesvirus 3}, partial (8%)
Length = 986
Score = 37.7 bits (86), Expect = 0.004
Identities = 26/76 (34%), Positives = 40/76 (52%)
Frame = -2
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASAT 69
+S SSS SSLSS+ S + P S S ++++ S S PSSS ++SP S+A
Sbjct: 748 ASSSSSSSSSSLSSSSTPSSSSSSPPSSSSSSSSSSSSSSSWSSPSSSSSSSPFSSAPPA 569
Query: 70 PSASSAIDFLSICHRL 85
P + + + L + L
Sbjct: 568 PPSGAFFELLLLLELL 521
>BQ750934 similar to PIR|T19673|T19 hypothetical protein C33B4.3 -
Caenorhabditis elegans, partial (1%)
Length = 627
Score = 37.0 bits (84), Expect = 0.008
Identities = 30/62 (48%), Positives = 35/62 (56%), Gaps = 1/62 (1%)
Frame = +2
Query: 14 SSSIPSSLSSTQLRFKSVTLSPHSP-SRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSA 72
SSSIP S SST L V + SP S +TT+ SSS SSS TTS S++S TP
Sbjct: 443 SSSIPES-SSTSLPPTLVPQNTTSPASSPSSITTSTTSSSSSSSSSTTSSSSSSSPTPPG 619
Query: 73 SS 74
S
Sbjct: 620 PS 625
Score = 27.7 bits (60), Expect = 4.6
Identities = 22/67 (32%), Positives = 32/67 (46%), Gaps = 2/67 (2%)
Frame = +2
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAA--S 67
+S +S+ PS + + + S S + TTA SSS P SS T+ PP+ +
Sbjct: 323 NSSVTSAAPSFTAEAETTSPVSSASSTSLETLDATTTAVPSSSIPESSSTSLPPTLVPQN 502
Query: 68 ATPSASS 74
T ASS
Sbjct: 503 TTSPASS 523
>TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {Arabidopsis
thaliana}, partial (39%)
Length = 883
Score = 36.6 bits (83), Expect = 0.010
Identities = 45/118 (38%), Positives = 62/118 (52%), Gaps = 9/118 (7%)
Frame = -1
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSS-PPSSS------VTTSP 62
SS SSS PSS SS+ S + S SPS + A+GSSS PSSS ++SP
Sbjct: 541 SSSSSSSSPSSSSSS-----SPSSSSSSPSSLPPAPLASGSSSGSPSSSPSSPE*SSSSP 377
Query: 63 PSAASATPSASSAIDFLSICHRLKTTKRAGWI--RKDVQDPESIADHMYRMSLMALIA 118
S++S T S+SS+ S+ L + AG++ R + ESIA + L ALI+
Sbjct: 376 SSSSSCTSSSSSSSSSSSV--SLLFSL*AGFLPRRCSKGNLESIAVNFGATELAALIS 209
Score = 31.6 bits (70), Expect = 0.32
Identities = 25/62 (40%), Positives = 36/62 (57%), Gaps = 6/62 (9%)
Frame = -1
Query: 20 SLSSTQLRFKSVTLSPHSPSRVFHMTTAAG------SSSPPSSSVTTSPPSAASATPSAS 73
SLS TQ++ T+ P S H AG SSS SSS ++SP S++S++PS+S
Sbjct: 649 SLSLTQMQ----TIEPISY*YNSHFLFFAGGWGVSSSSSSSSSSSSSSPSSSSSSSPSSS 482
Query: 74 SA 75
S+
Sbjct: 481 SS 476
Score = 26.9 bits (58), Expect = 7.9
Identities = 13/28 (46%), Positives = 24/28 (85%)
Frame = -1
Query: 45 TTAAGSSSPPSSSVTTSPPSAASATPSA 72
++++ SSS PSSS ++S PS++S++PS+
Sbjct: 544 SSSSSSSSSPSSS-SSSSPSSSSSSPSS 464
>TC90018 weakly similar to PIR|B84607|B84607 hypothetical protein At2g21950
[imported] - Arabidopsis thaliana, partial (55%)
Length = 965
Score = 35.8 bits (81), Expect = 0.017
Identities = 28/75 (37%), Positives = 39/75 (51%), Gaps = 6/75 (8%)
Frame = +1
Query: 8 LLSSLFSSSIPSSLSSTQLRFKSVTLSPHSP----SRVFHMTTAAGSS--SPPSSSVTTS 61
+LSSL SS P +LSS S +LS H+P S +TT + S PP + +
Sbjct: 139 ILSSLLRSSSPLALSSLPPNTSSTSLSAHAPPLSNSSPSTITTVSSLSLLFPP---LPSG 309
Query: 62 PPSAASATPSASSAI 76
PP+ +S T S SS +
Sbjct: 310 PPTLSSTTKSISSVV 354
>BG646467 similar to SP|O64668|PSNH Presenilin homolog. [Mouse-ear cress]
{Arabidopsis thaliana}, partial (49%)
Length = 796
Score = 35.4 bits (80), Expect = 0.022
Identities = 26/73 (35%), Positives = 39/73 (52%), Gaps = 9/73 (12%)
Frame = +1
Query: 16 SIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSS-------SVTTSPPSAA-- 66
S P +SST K+ PH S+ T + SSS PSS + TT+PPS+
Sbjct: 184 SPPPLISST---LKTPPTPPHKNSKAHFSTPSFSSSSSPSSPSFSSSFTTTTAPPSSVTM 354
Query: 67 SATPSASSAIDFL 79
SA+P +SS++ ++
Sbjct: 355 SASPPSSSSLPWV 393
>TC86287 similar to GP|23617108|dbj|BAC20790. contains ESTs AU056959(S21019)
AU056960(S21019)~similar to Arabidopsis thaliana
chromosome 5, partial (82%)
Length = 1302
Score = 35.0 bits (79), Expect = 0.029
Identities = 25/58 (43%), Positives = 31/58 (53%), Gaps = 5/58 (8%)
Frame = +3
Query: 36 HSPSRVFHM-----TTAAGSSSPPSSSVTTSPPSAASATPSASSAIDFLSICHRLKTT 88
+SPSR HM T SS+ SSS + SPPS S T S++S D + HR K T
Sbjct: 144 NSPSRGSHMGKILRETTRPSSNSSSSSSSPSPPSIPSTT-SSTSITDTIKGSHRFKIT 314
>TC82397 similar to GP|18377877|gb|AAL67124.1 AT3g53630/F4P12_330
{Arabidopsis thaliana}, partial (9%)
Length = 716
Score = 34.7 bits (78), Expect = 0.038
Identities = 23/62 (37%), Positives = 35/62 (56%), Gaps = 1/62 (1%)
Frame = -3
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGS-SSPPSSSVTTSPPSAASA 68
SS FS+ S S+ + + SPH+ +R+FH+ T S S+P S S T SP S+ +
Sbjct: 711 SSSFSNLTSSQPLSSPILPPTPIPSPHNNTRLFHLKTQIPSFSTPLSPSKTPSPSSSLLS 532
Query: 69 TP 70
+P
Sbjct: 531 SP 526
>AL368332 weakly similar to GP|21322711|e pherophorin-dz1 protein {Volvox
carteri f. nagariensis}, partial (10%)
Length = 478
Score = 34.7 bits (78), Expect = 0.038
Identities = 26/69 (37%), Positives = 34/69 (48%)
Frame = +3
Query: 15 SSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSASS 74
S IPSS SS+ + T+S S +TA S+SP S TT PP + T + S
Sbjct: 198 SFIPSSSSSSTQTPSTATISSIFSSTTTQPSTAI-STSPDCSFSTTRPPIPTTTTSTFSL 374
Query: 75 AIDFLSICH 83
+I S CH
Sbjct: 375 SIPPYSHCH 401
>TC88186 similar to GP|15293177|gb|AAK93699.1 unknown protein {Arabidopsis
thaliana}, partial (59%)
Length = 782
Score = 34.7 bits (78), Expect = 0.038
Identities = 20/47 (42%), Positives = 27/47 (56%)
Frame = -3
Query: 30 SVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSASSAI 76
S +S HSPS H ++ SS P SS S PS++ TPS SS++
Sbjct: 495 SFQISKHSPSSSSHSLLSSFSSPPYSSLSPLSTPSSSPYTPS*SSSL 355
>TC78873 similar to GP|21740530|emb|CAD41509. OSJNBa0029H02.25 {Oryza
sativa}, partial (7%)
Length = 623
Score = 34.3 bits (77), Expect = 0.050
Identities = 28/78 (35%), Positives = 39/78 (49%), Gaps = 3/78 (3%)
Frame = +3
Query: 7 VLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPS-- 64
+L+SS F+ S SS S KS +SP + S +S PS S SPPS
Sbjct: 153 LLVSSTFAQSPSSSPS------KSPAISPSAHSPAASPPAPVKNSPSPSPSAINSPPSPP 314
Query: 65 -AASATPSASSAIDFLSI 81
A+S +P+A+ A+ SI
Sbjct: 315 PASSGSPAAAPAVTPSSI 368
>BQ751225 similar to GP|18376350|em probable aldehyde dehydrogenase
{Neurospora crassa}, partial (31%)
Length = 568
Score = 34.3 bits (77), Expect = 0.050
Identities = 23/69 (33%), Positives = 35/69 (50%)
Frame = -1
Query: 16 SIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSASSA 75
S PS S + RF S SP +P V+ + +PPS+S T+S P+ + +T +
Sbjct: 364 SAPSPSSRMRRRFSS---SPTTPPTVWPPPFTPRT*TPPSASATSSRPAPSGSTATTCFT 194
Query: 76 IDFLSICHR 84
FLS+ R
Sbjct: 193 GRFLSVATR 167
>TC80941 weakly similar to PIR|T39903|T39903 serine-rich protein - fission
yeast (Schizosaccharomyces pombe), partial (15%)
Length = 557
Score = 33.9 bits (76), Expect = 0.065
Identities = 28/78 (35%), Positives = 38/78 (47%), Gaps = 6/78 (7%)
Frame = -3
Query: 9 LSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTS------P 62
LSS F SI SS +S+ + S SPS ++++ S S PSS +TS P
Sbjct: 351 LSSSFDLSIVSSSASSSSSVSASVFSFSSPSSFSSVSSSDFSLSSPSSFSSTSVFSSSLP 172
Query: 63 PSAASATPSASSAIDFLS 80
S +S PSAS + S
Sbjct: 171 SSFSSMLPSASFSASIFS 118
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.312 0.125 0.352
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,551,976
Number of Sequences: 36976
Number of extensions: 103978
Number of successful extensions: 1801
Number of sequences better than 10.0: 198
Number of HSP's better than 10.0 without gapping: 1364
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1606
length of query: 261
length of database: 9,014,727
effective HSP length: 94
effective length of query: 167
effective length of database: 5,538,983
effective search space: 925010161
effective search space used: 925010161
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0041.3


