

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0041.13
(130 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
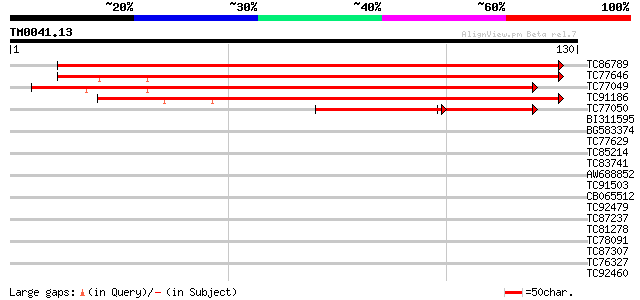
Score E
Sequences producing significant alignments: (bits) Value
TC86789 similar to PIR|A84594|A84594 hypothetical protein At2g20... 190 1e-49
TC77646 similar to GP|3941289|gb|AAC82326.1| similarity to SCAMP... 161 6e-41
TC77049 homologue to GP|3941289|gb|AAC82326.1| similarity to SCA... 154 1e-38
TC91186 similar to PIR|G86444|G86444 probable secretory carrier ... 124 8e-30
TC77050 homologue to GP|3941289|gb|AAC82326.1| similarity to SCA... 58 3e-14
BI311595 weakly similar to GP|21553662|gb| unknown {Arabidopsis ... 32 0.092
BG583374 30 0.20
TC77629 similar to GP|6648962|gb|AAF21309.1| seed maturation pro... 30 0.35
TC85214 similar to PIR|S26688|S26688 legumin K - garden pea, par... 29 0.60
TC83741 similar to GP|20453183|gb|AAM19832.1 At2g23390/F26B6.4 {... 28 1.0
AW688852 weakly similar to PIR|E71443|E714 probable DNA-binding ... 28 1.3
TC91503 similar to GP|20161013|dbj|BAB89946. putative ovule deve... 27 2.3
CB065512 similar to GP|3608154|gb| unknown protein {Arabidopsis ... 27 2.3
TC92479 similar to GP|9759160|dbj|BAB09716.1 contains similarity... 27 2.3
TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-inf... 27 3.0
TC81278 similar to GP|17979004|gb|AAL47462.1 At2g46180/T3F17.17 ... 27 3.0
TC78091 similar to GP|17104531|gb|AAL34154.1 unknown protein {Ar... 26 3.9
TC87307 weakly similar to PIR|T14329|T14329 dermal glycoprotein ... 26 3.9
TC76327 similar to GP|13359451|dbj|BAB33421. putative senescence... 26 3.9
TC92460 similar to GP|22202705|dbj|BAC07363. hypothetical protei... 26 5.0
>TC86789 similar to PIR|A84594|A84594 hypothetical protein At2g20840
[imported] - Arabidopsis thaliana, partial (93%)
Length = 1421
Score = 190 bits (483), Expect = 1e-49
Identities = 88/116 (75%), Positives = 104/116 (88%)
Frame = +3
Query: 12 NSGSVPLATSKLKPLPPEPYDRGATIDIPLDTTKDIKAKEKELQAREAELKRREQELKRK 71
N GSVP A S+L PLP EPYDR AT+DIPLD++KD+KAKEKELQARE+ELK+REQELKR+
Sbjct: 294 NPGSVPSAASRLSPLPHEPYDRNATVDIPLDSSKDVKAKEKELQARESELKKREQELKRR 473
Query: 72 EDAIARAGIVIEEKNWPPFFPIIHHEISKEIPLHLQRIQYVAFITWLGMLKFLFFS 127
EDAIARAGIVIEEKNWPPFFPIIHH+I EIP+HLQR+QY+AF TWLG++ L ++
Sbjct: 474 EDAIARAGIVIEEKNWPPFFPIIHHDIGTEIPIHLQRMQYIAFSTWLGLVLCLLWN 641
>TC77646 similar to GP|3941289|gb|AAC82326.1| similarity to SCAMP37 {Pisum
sativum}, partial (89%)
Length = 1448
Score = 161 bits (408), Expect = 6e-41
Identities = 78/122 (63%), Positives = 101/122 (81%), Gaps = 6/122 (4%)
Frame = +1
Query: 12 NSGSVPLA-TSKLKPLPPEP-----YDRGATIDIPLDTTKDIKAKEKELQAREAELKRRE 65
N GSVP A S+L PL PEP Y GAT+DIPLDT+ D+K +EKELQ++EA+L+RRE
Sbjct: 328 NPGSVPPAKNSRLSPLEPEPADYNNYGFGATVDIPLDTSTDLKKREKELQSKEADLRRRE 507
Query: 66 QELKRKEDAIARAGIVIEEKNWPPFFPIIHHEISKEIPLHLQRIQYVAFITWLGMLKFLF 125
QE++RKE+A ARAGIV+EEKNWPPFFPIIHH+I+ EIP+HLQ++QYVAF T+LG++ L
Sbjct: 508 QEVRRKEEAAARAGIVLEEKNWPPFFPIIHHDIANEIPVHLQKLQYVAFTTYLGLVACLL 687
Query: 126 FS 127
++
Sbjct: 688 WN 693
>TC77049 homologue to GP|3941289|gb|AAC82326.1| similarity to SCAMP37 {Pisum
sativum}, complete
Length = 1358
Score = 154 bits (388), Expect = 1e-38
Identities = 75/121 (61%), Positives = 96/121 (78%), Gaps = 5/121 (4%)
Frame = +1
Query: 6 EMEPARNSGSV-PLATSKLKPLPPEP----YDRGATIDIPLDTTKDIKAKEKELQAREAE 60
++ P NSG V P S+ PL PE Y G T+DIPLD + D+K KE+ELQA+EAE
Sbjct: 274 QVNPFSNSGGVAPATNSRPAPLNPERAGYNYGFGQTVDIPLDASTDVKKKERELQAKEAE 453
Query: 61 LKRREQELKRKEDAIARAGIVIEEKNWPPFFPIIHHEISKEIPLHLQRIQYVAFITWLGM 120
L++REQE++RKE+AI+RAGIVIEEKNWPPFFPIIHH+I+ EIP+HLQR+QYVAF + LG+
Sbjct: 454 LRKREQEVRRKEEAISRAGIVIEEKNWPPFFPIIHHDIANEIPVHLQRLQYVAFFSLLGL 633
Query: 121 L 121
+
Sbjct: 634 V 636
>TC91186 similar to PIR|G86444|G86444 probable secretory carrier membrane
protein [imported] - Arabidopsis thaliana, partial (56%)
Length = 592
Score = 124 bits (312), Expect = 8e-30
Identities = 58/112 (51%), Positives = 85/112 (75%), Gaps = 5/112 (4%)
Frame = +3
Query: 21 SKLKPLPPEPYDRG----ATIDIPLDTTK-DIKAKEKELQAREAELKRREQELKRKEDAI 75
S++ + EP G AT+DIPL+T+ D K K +EL EA+LKRRE+++KR+ED++
Sbjct: 153 SRVPSVASEPLGFGQRHDATVDIPLETSNGDSKKKSQELAVWEADLKRREKDIKRREDSV 332
Query: 76 ARAGIVIEEKNWPPFFPIIHHEISKEIPLHLQRIQYVAFITWLGMLKFLFFS 127
A+AG+ +++KNWPPFFPIIHH+I+ EIP+H QR+QY AF +WLG++ L F+
Sbjct: 333 AKAGVPVDDKNWPPFFPIIHHDIANEIPVHAQRLQYSAFASWLGIVLCLVFN 488
>TC77050 homologue to GP|3941289|gb|AAC82326.1| similarity to SCAMP37 {Pisum
sativum}, partial (51%)
Length = 459
Score = 57.8 bits (138), Expect(2) = 3e-14
Identities = 24/30 (80%), Positives = 27/30 (90%)
Frame = +3
Query: 71 KEDAIARAGIVIEEKNWPPFFPIIHHEISK 100
KE+AI+RAGIVIEEKNWPPFFPIIHH +
Sbjct: 3 KEEAISRAGIVIEEKNWPPFFPIIHHRYXR 92
Score = 35.4 bits (80), Expect(2) = 3e-14
Identities = 14/23 (60%), Positives = 20/23 (86%)
Frame = +2
Query: 99 SKEIPLHLQRIQYVAFITWLGML 121
+ EIP+HLQR+QYVAF + LG++
Sbjct: 89 ANEIPVHLQRLQYVAFFSLLGLV 157
>BI311595 weakly similar to GP|21553662|gb| unknown {Arabidopsis thaliana},
partial (21%)
Length = 320
Score = 31.6 bits (70), Expect = 0.092
Identities = 29/87 (33%), Positives = 43/87 (49%), Gaps = 7/87 (8%)
Frame = +2
Query: 6 EMEPARNSGS-VPLATSKLKPLPPEPYDRGATIDIPLDTTKDI------KAKEKELQARE 58
+MEP S VPL KP P +P T ++ + K + +AK+ + +A
Sbjct: 26 DMEPNHERESTVPLP----KPEPSQP-----TSELSKNAKKKLARQERWEAKKADKKAAA 178
Query: 59 AELKRREQELKRKEDAIARAGIVIEEK 85
E K++E E KRKE + AGI EE+
Sbjct: 179 KEQKKKETERKRKEWEESIAGITEEER 259
>BG583374
Length = 815
Score = 30.4 bits (67), Expect = 0.20
Identities = 16/69 (23%), Positives = 31/69 (44%)
Frame = +2
Query: 17 PLATSKLKPLPPEPYDRGATIDIPLDTTKDIKAKEKELQAREAELKRREQELKRKEDAIA 76
PL+TSK K E I P+ T+ + +E + + + ++ EDA+
Sbjct: 395 PLSTSKHKTSSQEYGGGEMAISSPISITRPLTTQEVHVNNLQETSNTNDDQVSLNEDAVV 574
Query: 77 RAGIVIEEK 85
+ ++EE+
Sbjct: 575 KVTSIVEEQ 601
>TC77629 similar to GP|6648962|gb|AAF21309.1| seed maturation protein PM23
{Glycine max}, partial (76%)
Length = 1741
Score = 29.6 bits (65), Expect = 0.35
Identities = 18/53 (33%), Positives = 29/53 (53%), Gaps = 4/53 (7%)
Frame = +3
Query: 38 DIPLDTTKDIKAKEKE----LQAREAELKRREQELKRKEDAIARAGIVIEEKN 86
D+ + I A+ +E LQ+R + +K+ +E+KRK A+ V EEKN
Sbjct: 669 DVNIQDLGQISAEAQEYISSLQSRLSSIKKELREVKRKSAALQMQQFVGEEKN 827
>TC85214 similar to PIR|S26688|S26688 legumin K - garden pea, partial (94%)
Length = 1831
Score = 28.9 bits (63), Expect = 0.60
Identities = 18/61 (29%), Positives = 26/61 (42%)
Frame = +3
Query: 50 KEKELQAREAELKRREQELKRKEDAIARAGIVIEEKNWPPFFPIIHHEISKEIPLHLQRI 109
+E+E + R L RRE + +E G EE+ W P P E + P H +
Sbjct: 888 EEEEDEGRSRHLLRREHSQEERERDPRHPGHSQEEREWDPRHPGHSQEERERDPRHPRHS 1067
Query: 110 Q 110
Q
Sbjct: 1068Q 1070
>TC83741 similar to GP|20453183|gb|AAM19832.1 At2g23390/F26B6.4 {Arabidopsis
thaliana}, partial (25%)
Length = 690
Score = 28.1 bits (61), Expect = 1.0
Identities = 16/53 (30%), Positives = 25/53 (46%), Gaps = 6/53 (11%)
Frame = +1
Query: 82 IEEKNWPPFFPI---IHHEISKEIPLHLQRIQYVAFI---TWLGMLKFLFFSN 128
++EK W P I IH+ I +PL+L+ Y F+ +W + SN
Sbjct: 424 VKEKGWTPHHIIAKDIHNHILAVVPLYLKTHSYGEFVFDHSWANAYSHFYGSN 582
>AW688852 weakly similar to PIR|E71443|E714 probable DNA-binding protein -
Arabidopsis thaliana, partial (9%)
Length = 536
Score = 27.7 bits (60), Expect = 1.3
Identities = 13/50 (26%), Positives = 28/50 (56%)
Frame = +2
Query: 23 LKPLPPEPYDRGATIDIPLDTTKDIKAKEKELQAREAELKRREQELKRKE 72
+ P+PP P+ A + ++ ++E E +AR+A+ +R+ + +R E
Sbjct: 371 MPPIPPPPHRDLAEFSMGMNVPPPAMSRE-EFEARKADARRKRENERRVE 517
>TC91503 similar to GP|20161013|dbj|BAB89946. putative ovule development
protein aintegumenta-like protein {Oryza sativa
(japonica cultivar-group), partial (27%)
Length = 1057
Score = 26.9 bits (58), Expect = 2.3
Identities = 18/47 (38%), Positives = 23/47 (48%), Gaps = 4/47 (8%)
Frame = +2
Query: 71 KEDAIARAGIVIEEKNWPPF----FPIIHHEISKEIPLHLQRIQYVA 113
KE+ ARA + K W FPI H+E E H+ R +YVA
Sbjct: 56 KEEKAARAYDLAALKYWGTTTTTNFPISHYEKEVEEMKHMTRQEYVA 196
>CB065512 similar to GP|3608154|gb| unknown protein {Arabidopsis thaliana},
partial (34%)
Length = 852
Score = 26.9 bits (58), Expect = 2.3
Identities = 18/41 (43%), Positives = 29/41 (69%)
Frame = +1
Query: 45 KDIKAKEKELQAREAELKRREQELKRKEDAIARAGIVIEEK 85
+++ A+ K+ Q +EAE K +E ELK +EDA+A +V EE+
Sbjct: 388 REVSAQNKQRQ-KEAEAKWKE-ELKAQEDALA---LVDEER 495
>TC92479 similar to GP|9759160|dbj|BAB09716.1 contains similarity to myosin
heavy chain~gene_id:MEE6.21 {Arabidopsis thaliana},
partial (4%)
Length = 844
Score = 26.9 bits (58), Expect = 2.3
Identities = 18/47 (38%), Positives = 26/47 (55%), Gaps = 4/47 (8%)
Frame = +2
Query: 43 TTKDIKAKEKELQAREAELKRREQELKRK----EDAIARAGIVIEEK 85
TT +K KEKELQ+R EL+ + +E + ED ++ I EK
Sbjct: 257 TTSSMK-KEKELQSRIVELENKVEEFNQNVTLHEDRSIKSSNEISEK 394
>TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-infected
erythrocyte surface antigen {Plasmodium falciparum},
partial (2%)
Length = 2007
Score = 26.6 bits (57), Expect = 3.0
Identities = 11/39 (28%), Positives = 23/39 (58%)
Frame = +1
Query: 48 KAKEKELQAREAELKRREQELKRKEDAIARAGIVIEEKN 86
K++++ + ++ E ++E ELK+ E G + EEK+
Sbjct: 232 KSQQENEENKDEEKSQQENELKKNEGGEKETGEITEEKS 348
>TC81278 similar to GP|17979004|gb|AAL47462.1 At2g46180/T3F17.17
{Arabidopsis thaliana}, partial (10%)
Length = 857
Score = 26.6 bits (57), Expect = 3.0
Identities = 19/62 (30%), Positives = 27/62 (42%), Gaps = 4/62 (6%)
Frame = +1
Query: 20 TSKLKPLPPEPYDRGATIDIPLDTTKDIKAKEKELQAREAELKRREQE----LKRKEDAI 75
+++ +P P G + +A+ K LQA EAE+K LK KED I
Sbjct: 298 STRSNSIPRSPIPNGIADHPYSSEIEQYRAEIKRLQASEAEIKALSVNYAALLKEKEDHI 477
Query: 76 AR 77
R
Sbjct: 478 IR 483
>TC78091 similar to GP|17104531|gb|AAL34154.1 unknown protein {Arabidopsis
thaliana}, partial (52%)
Length = 1863
Score = 26.2 bits (56), Expect = 3.9
Identities = 13/29 (44%), Positives = 18/29 (61%)
Frame = +2
Query: 17 PLATSKLKPLPPEPYDRGATIDIPLDTTK 45
P+ TS+ PLPP P+D A + IP +K
Sbjct: 1322 PMKTSE--PLPPAPWDTQAPVVIPPPPSK 1402
>TC87307 weakly similar to PIR|T14329|T14329 dermal glycoprotein precursor
extracellular - carrot (fragment), partial (18%)
Length = 1500
Score = 26.2 bits (56), Expect = 3.9
Identities = 11/28 (39%), Positives = 17/28 (60%)
Frame = +1
Query: 100 KEIPLHLQRIQYVAFITWLGMLKFLFFS 127
K+IPL +Q++ F++ L F FFS
Sbjct: 64 KKIPLSFLLLQWLLFLSLFSFLSFSFFS 147
>TC76327 similar to GP|13359451|dbj|BAB33421. putative senescence-associated
protein {Pisum sativum}, partial (64%)
Length = 868
Score = 26.2 bits (56), Expect = 3.9
Identities = 12/30 (40%), Positives = 18/30 (60%)
Frame = +2
Query: 83 EEKNWPPFFPIIHHEISKEIPLHLQRIQYV 112
E +N F+P + HEIS I L L ++Y+
Sbjct: 461 ENQNQMSFYPFVLHEISVLIELILGHLRYL 550
>TC92460 similar to GP|22202705|dbj|BAC07363. hypothetical protein {Oryza
sativa (japonica cultivar-group)}, partial (9%)
Length = 795
Score = 25.8 bits (55), Expect = 5.0
Identities = 13/60 (21%), Positives = 26/60 (42%)
Frame = +3
Query: 14 GSVPLATSKLKPLPPEPYDRGATIDIPLDTTKDIKAKEKELQAREAELKRREQELKRKED 73
G V SK KP+PP R ++ P+ + +E++ ++ + + +ED
Sbjct: 27 GDVSSIKSKTKPIPPSSASRLSSKPSPVQNGHRQDSSHREVEPPRPTKRKSDHSERERED 206
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.138 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,840,447
Number of Sequences: 36976
Number of extensions: 45211
Number of successful extensions: 390
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 386
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 388
length of query: 130
length of database: 9,014,727
effective HSP length: 85
effective length of query: 45
effective length of database: 5,871,767
effective search space: 264229515
effective search space used: 264229515
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 53 (25.0 bits)
Lotus: description of TM0041.13


