

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0040.4
(457 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
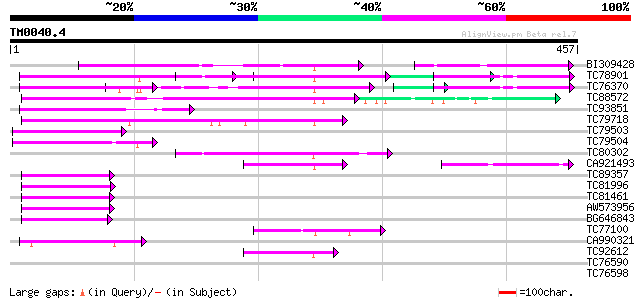
Score E
Sequences producing significant alignments: (bits) Value
BI309428 weakly similar to GP|21734794|gb| reduced vernalization... 100 1e-21
TC78901 similar to GP|21734794|gb|AAM76972.1 reduced vernalizati... 95 6e-20
TC76370 similar to GP|21734794|gb|AAM76972.1 reduced vernalizati... 90 2e-18
TC88572 weakly similar to GP|20145851|emb|CAD29616. auxin respon... 84 1e-16
TC93851 weakly similar to GP|21734794|gb|AAM76972.1 reduced vern... 81 1e-15
TC79718 80 2e-15
TC79503 weakly similar to GP|21734794|gb|AAM76972.1 reduced vern... 73 2e-13
TC79504 weakly similar to GP|21734794|gb|AAM76972.1 reduced vern... 70 2e-12
TC80302 57 2e-08
CA921493 51 8e-07
TC89357 similar to GP|20145851|emb|CAD29616. auxin response fact... 51 8e-07
TC81996 similar to GP|20145851|emb|CAD29616. auxin response fact... 50 2e-06
TC81461 49 3e-06
AW573956 similar to GP|20145851|emb auxin response factor 36 {Ar... 48 7e-06
BG646843 similar to GP|20145851|emb auxin response factor 36 {Ar... 48 9e-06
TC77100 42 5e-04
CA990321 weakly similar to PIR|S03170|S03 homeotic protein cut -... 42 5e-04
TC92612 41 8e-04
TC76590 similar to GP|14532680|gb|AAK64141.1 unknown protein {Ar... 39 0.003
TC76598 similar to GP|14532680|gb|AAK64141.1 unknown protein {Ar... 39 0.005
>BI309428 weakly similar to GP|21734794|gb| reduced vernalization response 1
{Arabidopsis thaliana}, partial (6%)
Length = 710
Score = 100 bits (249), Expect = 1e-21
Identities = 74/239 (30%), Positives = 115/239 (47%), Gaps = 9/239 (3%)
Frame = +1
Query: 56 FAKYYSLDHGHMVLFEYKDTSHFGVHIFDKSTLEIRY-PSHENQDEEHNPDQIDDD-SVE 113
F + YSL HG +V+FEYK T F V I K+ LEI Y S ++ DE N D DDD SVE
Sbjct: 4 FTENYSLGHGCLVVFEYKGTFKFDVVILGKNALEIDYDTSCDSDDENDNVDHSDDDESVE 183
Query: 114 ILDKIPPCKKTKLKSPMSCPKPRKKLRTSTSEDVGESPELHNLPKQVKIEDDAGRTTECL 173
ILD+ KK + +SP +P KK++ + + + N PK V+ ++
Sbjct: 184 ILDEWLNRKKARQRSPFVSSRPHKKVQGDDEKTTKRTSSM-NWPKDVRAQE--------- 333
Query: 174 EGEDLTSKITEALNRARTFQSKFPSFMTVMKPAYVNGYSLHIPSFFAKKYLKNETDVLLQ 233
A+ F S P F ++KP + ++ +P K ++N+ ++
Sbjct: 334 --------------VAQNFISCNPFFTILIKPINLVEHTRSVPDL--KGVIENKDTNVML 465
Query: 234 VLDGRTWSVSF-------NLGKFNAGWKKFTSVNNLKVGDVCLFELNKSVPLSLKVLIF 285
++ R+W+V N + +AGW F + LK GDVC+FEL L K+ ++
Sbjct: 466 LIGKRSWNVKLLRSYEGKNGRRLSAGWYLFARESGLKSGDVCVFELINKKDLVFKIHVY 642
Score = 76.3 bits (186), Expect = 2e-14
Identities = 52/129 (40%), Positives = 69/129 (53%), Gaps = 1/129 (0%)
Frame = +1
Query: 327 ETIKRTSKLISPCPLKNAALEEANKFNSNNPFFIVNIAPDKNRDYRPRVPKLFIRKHFND 386
+T KRTS + P ++ A E A F S NPFF + I P ++ VP L
Sbjct: 277 KTTKRTSSMNWPKDVR--AQEVAQNFISCNPFFTILIKPINLVEHTRSVPDLK-----GV 435
Query: 387 IQQSEQTVILQCEKKLCPVKLT-CYPKKGPSRLSKGWVEFAKKSKLEVGDACVFELINKE 445
I+ + V+L K+ VKL Y K RLS GW FA++S L+ GD CVFELINK+
Sbjct: 436 IENKDTNVMLLIGKRSWNVKLLRSYEGKNGRRLSAGWYLFARESGLKSGDVCVFELINKK 615
Query: 446 DPVLDVHIY 454
D V +H+Y
Sbjct: 616 DLVFKIHVY 642
>TC78901 similar to GP|21734794|gb|AAM76972.1 reduced vernalization response
1 {Arabidopsis thaliana}, partial (37%)
Length = 1760
Score = 94.7 bits (234), Expect = 6e-20
Identities = 54/184 (29%), Positives = 86/184 (46%), Gaps = 9/184 (4%)
Frame = +2
Query: 9 RIPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNGWKDFAKYYSLDHGHMV 68
R+P++F ++YGGD++ L PDG+ W++ K D WF +GW +F + YS+ G+++
Sbjct: 266 RLPDDFMRKYGGDISPTVTLTVPDGSVWRVIMKKVDNKFWFLDGWNEFVQNYSISTGYLL 445
Query: 69 LFEYKDTSHFGVHIFDKSTLEIRYPSHENQDEEHN---------PDQIDDDSVEILDKIP 119
+F+Y+ SHF V+IF T EI Y S + E + + D+DSVEI++ P
Sbjct: 446 VFKYEGKSHFTVNIFSLPTSEINYQSPAQRSNEASLFGKRLTIFEEMEDEDSVEIMESSP 625
Query: 120 PCKKTKLKSPMSCPKPRKKLRTSTSEDVGESPELHNLPKQVKIEDDAGRTTECLEGEDLT 179
L + KL S L N K I G T +
Sbjct: 626 TKLTPSLLQNKAVSGSADKLTPGKSRPPPALQNLFNGSKLNSINWGEGGNTPSRNDNSVD 805
Query: 180 SKIT 183
+++T
Sbjct: 806 NQLT 817
Score = 81.3 bits (199), Expect = 7e-16
Identities = 55/181 (30%), Positives = 91/181 (49%), Gaps = 7/181 (3%)
Frame = +2
Query: 134 KPRKKLRTSTSEDVGESPELHNLPKQVKIEDDAGRTTECLEGEDLTSKITEALNRARTFQ 193
K +K DV E P H +V++ + + + + +A+N A+TF+
Sbjct: 899 KVKKTAVKKRKSDVQEPPSEHE--DEVEMRNRFYESASARKRTATAEEREKAINAAKTFE 1072
Query: 194 SKFPSFMTVMKPAYV-NGYSLHIPSFFAKKYLKNETDVL-LQVLDGRTWSVSF----NLG 247
P V++P+Y+ G +++PS FA+K L + ++ LQ+ DGR W V
Sbjct: 1073 PSNPFCRVVLRPSYLYRGCIMYLPSCFAEKNLNGVSGIIKLQISDGRQWPVRCLYRGGRA 1252
Query: 248 KFNAGWKKFTSVNNLKVGDVCLFELNKSVPLSLKVLIFPLAEEPHSL-PRSLVQGDRVNH 306
K + GW +F+ NNL GDVC+FEL + + L+V +F + E+ L P+ L Q VN
Sbjct: 1253 KLSQGWFEFSLENNLGEGDVCVFELVATKEVVLQVTVFRITEDEGPLSPQPLQQNQHVNP 1432
Query: 307 I 307
+
Sbjct: 1433 V 1435
Score = 55.8 bits (133), Expect = 3e-08
Identities = 36/115 (31%), Positives = 59/115 (51%), Gaps = 1/115 (0%)
Frame = +2
Query: 342 KNAALEEANKFNSNNPFFIVNIAPDK-NRDYRPRVPKLFIRKHFNDIQQSEQTVILQCEK 400
+ A+ A F +NPF V + P R +P F K+ N + + I +
Sbjct: 1037 REKAINAAKTFEPSNPFCRVVLRPSYLYRGCIMYLPSCFAEKNLNGVSGIIKLQI--SDG 1210
Query: 401 KLCPVKLTCYPKKGPSRLSKGWVEFAKKSKLEVGDACVFELINKEDPVLDVHIYR 455
+ PV+ C + G ++LS+GW EF+ ++ L GD CVFEL+ ++ VL V ++R
Sbjct: 1211 RQWPVR--CLYRGGRAKLSQGWFEFSLENNLGEGDVCVFELVATKEVVLQVTVFR 1369
Score = 42.0 bits (97), Expect = 5e-04
Identities = 46/201 (22%), Positives = 78/201 (37%), Gaps = 6/201 (2%)
Frame = +2
Query: 197 PSFMTVMKPAYVNGYSLHIPSFFAKKYLKN-ETDVLLQVLDGRTWSVSF----NLGKFNA 251
PSF ++ P+ + L +P F +KY + V L V DG W V N F
Sbjct: 215 PSFHKLVLPSTLQAKQLRLPDDFMRKYGGDISPTVTLTVPDGSVWRVIMKKVDNKFWFLD 394
Query: 252 GWKKFTSVNNLKVGDVCLFELNKSVPLSLKVLIFPLAEEPHSLPRSLVQGDRVNHINYGR 311
GW +F ++ G + +F+ ++ + P +E + P R N +
Sbjct: 395 GWNEFVQNYSISTGYLLVFKYEGKSHFTVNIFSLPTSEINYQSP-----AQRSNEASLFG 559
Query: 312 FRYTESQNVLTSKCAETIKRTSKLISPCPLKNAALE-EANKFNSNNPFFIVNIAPDKNRD 370
R T + + E ++ + ++P L+N A+ A+K + P K+R
Sbjct: 560 KRLTIFEEMEDEDSVEIMESSPTKLTPSLLQNKAVSGSADK-----------LTPGKSRP 706
Query: 371 YRPRVPKLFIRKHFNDIQQSE 391
P + LF N I E
Sbjct: 707 -PPALQNLFNGSKLNSINWGE 766
>TC76370 similar to GP|21734794|gb|AAM76972.1 reduced vernalization response
1 {Arabidopsis thaliana}, partial (36%)
Length = 1840
Score = 90.1 bits (222), Expect = 2e-18
Identities = 46/120 (38%), Positives = 68/120 (56%), Gaps = 9/120 (7%)
Frame = +1
Query: 9 RIPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNGWKDFAKYYSLDHGHMV 68
RIP+NF ++YG ++ L PDGT W++ K D +WF +GW+DF + Y++ G+ +
Sbjct: 190 RIPDNFLRKYGAQLSTIATLTVPDGTVWRLGLKKVDNRIWFVDGWQDFVQRYAIGIGYFL 369
Query: 69 LFEYKDTSHFGVHIFDKSTLEIRYPSHENQDEE------HNP---DQIDDDSVEILDKIP 119
+F Y+ S+F VHIF+ ST E+ Y S E ++P D D DS E LD P
Sbjct: 370 VFTYEGNSNFIVHIFNMSTAELNYQSAMRSRTEGPCYANYHPIFEDIEDIDSFEFLDSSP 549
Score = 77.0 bits (188), Expect(2) = 9e-15
Identities = 66/225 (29%), Positives = 105/225 (46%), Gaps = 8/225 (3%)
Frame = +1
Query: 78 FGVHIFDKST--LEIRYPSHENQDEEHNPDQIDDDSVEILDKIPPCKKTKLKSPMSCPKP 135
F + F KST L++RYP N++ +N D KKT K S P
Sbjct: 751 FNANEFKKSTEGLKLRYP---NEEGVNNTDT---------------KKTSRKKRKSDPSA 876
Query: 136 RKKLRTSTSEDVGESPELHNLPKQVKIEDDAGRTTECLEGEDLTSKITEALNRARTFQSK 195
++ T+ +++ E + K RT E E A+N ++TF+
Sbjct: 877 QEA--TAENDEEAERYRFYESASARK------RTVTAEERE-------RAINESKTFEPT 1011
Query: 196 FPSFMTVMKPAYV-NGYSLHIPSFFAKKYLKNETDVL-LQVLDGRTWSVSF----NLGKF 249
P V++P+Y+ G +++PS FA+K L + + LQ+ DGR W V K
Sbjct: 1012NPFCRVVLRPSYLYRGCIMYLPSCFAEKNLNGVSGFIKLQISDGRQWPVRCLYKGGRAKL 1191
Query: 250 NAGWKKFTSVNNLKVGDVCLFELNKSVPLSLKVLIFPLAEEPHSL 294
+ GW +FT NNL GD+C+FEL ++ + L+V +F + E+ L
Sbjct: 1192SQGWYEFTLENNLGEGDICVFELLRTREVVLQVTLFRVKEDDSGL 1326
Score = 56.6 bits (135), Expect = 2e-08
Identities = 37/115 (32%), Positives = 57/115 (49%), Gaps = 1/115 (0%)
Frame = +1
Query: 342 KNAALEEANKFNSNNPFFIVNIAPDK-NRDYRPRVPKLFIRKHFNDIQQSEQTVILQCEK 400
+ A+ E+ F NPF V + P R +P F K+ N + + I +
Sbjct: 970 RERAINESKTFEPTNPFCRVVLRPSYLYRGCIMYLPSCFAEKNLNGVSGFIKLQI--SDG 1143
Query: 401 KLCPVKLTCYPKKGPSRLSKGWVEFAKKSKLEVGDACVFELINKEDPVLDVHIYR 455
+ PV+ C K G ++LS+GW EF ++ L GD CVFEL+ + VL V ++R
Sbjct: 1144 RQWPVR--CLYKGGRAKLSQGWYEFTLENNLGEGDICVFELLRTREVVLQVTLFR 1302
Score = 31.6 bits (70), Expect = 0.67
Identities = 23/79 (29%), Positives = 34/79 (42%), Gaps = 5/79 (6%)
Frame = +1
Query: 197 PSFMTVMKPAYVNGYSLHIPSFFAKKY-LKNETDVLLQVLDGRTWSVSF----NLGKFNA 251
PSF ++ P+ + L IP F +KY + T L V DG W + N F
Sbjct: 139 PSFHKLILPSSLQTRQLRIPDNFLRKYGAQLSTIATLTVPDGTVWRLGLKKVDNRIWFVD 318
Query: 252 GWKKFTSVNNLKVGDVCLF 270
GW+ F + +G +F
Sbjct: 319 GWQDFVQRYAIGIGYFLVF 375
Score = 28.1 bits (61), Expect = 7.4
Identities = 23/113 (20%), Positives = 40/113 (35%), Gaps = 12/113 (10%)
Frame = +1
Query: 2 LMESFVQRIPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNGWKDFAKYYS 61
L + +P+ F ++ ++ L+ DG W + G GW +F +
Sbjct: 1048 LYRGCIMYLPSCFAEKNLNGVSGFIKLQISDGRQWPVRCLYKGGRAKLSQGWYEFTLENN 1227
Query: 62 LDHGHMVLFEYKDTSHFGVHI------------FDKSTLEIRYPSHENQDEEH 102
L G + +FE T + + F+ STL + SH H
Sbjct: 1228 LGEGDICVFELLRTREVVLQVTLFRVKEDDSGLFNPSTLPSQNVSHAKMLNPH 1386
Score = 20.8 bits (42), Expect(2) = 9e-15
Identities = 13/45 (28%), Positives = 18/45 (39%)
Frame = +3
Query: 310 GRFRYTESQNVLTSKCAETIKRTSKLISPCPLKNAALEEANKFNS 354
GR R + KC S L +PC L + + A + NS
Sbjct: 1311 GRLRVIQPFYTAEPKCEPCKNVESSLAAPC*LNQIS*KLAGRDNS 1445
>TC88572 weakly similar to GP|20145851|emb|CAD29616. auxin response factor
36 {Arabidopsis thaliana}, partial (21%)
Length = 1755
Score = 83.6 bits (205), Expect = 1e-16
Identities = 76/283 (26%), Positives = 119/283 (41%), Gaps = 10/283 (3%)
Frame = +2
Query: 10 IPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNGWKDFAKYYSLDHGHMVL 69
+P F+ + LK P G W I + D V+F +GWK F K +SL ++
Sbjct: 608 LPKTFSDNLKKKLPENVTLKGPSGVVWNIGLTTRDDIVYFVDGWKRFIKDHSLKQNDFLV 787
Query: 70 FEYKDTSHFGVHIFDKSTLEIRYPSHENQDEEHNPDQIDDDSVEILDKIPPCKKTKLKSP 129
F+Y S F V IFD + + S+ + QI+ K K
Sbjct: 788 FKYNGKSLFEVLIFDGDSFCEKAASYFVG--KCGNAQIEQGG------------RKAKDT 925
Query: 130 MSCPKPRKKLRTST-SEDVGESPELHNLPKQVKIEDDAGRTTECLEGEDLTSKITEALNR 188
P + L + S+ G+ + + + ++ GR + S
Sbjct: 926 NKSVSPEQFLADAVPSQTNGKRTKKRPVNEVTPLQTKRGRRPKAEAALSKLSSSHAEKKI 1105
Query: 189 ARTFQSKFPSFMTVMKPAYVNGYS-LHIPSFFAKKYLKN-ETDVLLQVLDGRTWSV-SFN 245
A +F S FP F+ ++K V+G L++P F+K +L N + ++L L G W+V S
Sbjct: 1106AESFTSSFPYFVKMIKTFNVDGPRILNVPHQFSKAHLPNRKIKIILHNLKGEQWTVNSVP 1285
Query: 246 LGKFNA------GWKKFTSVNNLKVGDVCLFELNKSVPLSLKV 282
+ N GW F NN+KVGDVC+FEL L ++V
Sbjct: 1286TTRVNTDHTLCGGWVNFVRGNNIKVGDVCIFELIHECELRVRV 1414
Score = 53.1 bits (126), Expect = 2e-07
Identities = 103/476 (21%), Positives = 173/476 (35%), Gaps = 41/476 (8%)
Frame = +2
Query: 10 IPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNGWKDFAKYYSLDHGHMVL 69
+P F+ + LK P G W I + D ++ +GW+ F K +SL ++
Sbjct: 137 LPKTFSDNLKNKLPANVTLKGPSGVVWDIGLTTRDDTIYSTDGWQQFVKDHSLKQNDFLV 316
Query: 70 FEYKDTSHFGVHIFDKSTLEIRYPSHENQDEEHNPDQIDDDSVEILDKIPPCKKTKLKSP 129
F+Y S F V IF + C+K
Sbjct: 317 FKYNGESLFEVLIFHGESF--------------------------------CEKAASYFI 400
Query: 130 MSCPKPRKKLRTSTSEDVGESPELHNLPKQVKIEDDAGRTTECLEGEDLTSKITEALNRA 189
+ + R ++D S E N P +E G + E ED+ + ++
Sbjct: 401 LERGQAHTGQRGRKAKDTNTSVEEVNTPSNGSVE--CGLPEKSWE-EDIYWTHFQFIHFT 571
Query: 190 RTFQSKFPSFMTVMKPAYVNGYSLHIPSFFAKKYLKN-ETDVLLQVLDGRTWSVSF---- 244
+ ++ F L +P F+ K +V L+ G W++
Sbjct: 572 QFLRADFEQ-------------QLALPKTFSDNLKKKLPENVTLKGPSGVVWNIGLTTRD 712
Query: 245 NLGKFNAGWKKFTSVNNLKVGDVCLFELN-KSVPLSLKVLIF---PLAEEPHSL------ 294
++ F GWK+F ++LK D +F+ N KS+ +VLIF E+ S
Sbjct: 713 DIVYFVDGWKRFIKDHSLKQNDFLVFKYNGKSL---FEVLIFDGDSFCEKAASYFVGKCG 883
Query: 295 PRSLVQG-----DRVNHINYGRFRYTESQNVLTSKCAETIKRTSKLISPC-------PLK 342
+ QG D ++ +F + K T KR ++P P
Sbjct: 884 NAQIEQGGRKAKDTNKSVSPEQFLADAVPSQTNGK--RTKKRPVNEVTPLQTKRGRRPKA 1057
Query: 343 NAALEE----------ANKFNSNNPFFIVNIAPDKNRDYRPR---VPKLFIRKHFNDIQQ 389
AAL + A F S+ P+F V + N D PR VP F + H + +
Sbjct: 1058EAALSKLSSSHAEKKIAESFTSSFPYF-VKMIKTFNVD-GPRILNVPHQFSKAHLPN--R 1225
Query: 390 SEQTVILQCEKKLCPVKLTCYPKKGPSR-LSKGWVEFAKKSKLEVGDACVFELINK 444
+ ++ + + V + L GWV F + + ++VGD C+FELI++
Sbjct: 1226KIKIILHNLKGEQWTVNSVPTTRVNTDHTLCGGWVNFVRGNNIKVGDVCIFELIHE 1393
>TC93851 weakly similar to GP|21734794|gb|AAM76972.1 reduced vernalization
response 1 {Arabidopsis thaliana}, partial (9%)
Length = 710
Score = 80.9 bits (198), Expect = 1e-15
Identities = 48/142 (33%), Positives = 70/142 (48%), Gaps = 1/142 (0%)
Frame = +1
Query: 9 RIPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNG-WKDFAKYYSLDHGHM 67
RIP NF +G ++ N + PDG DW++ K DV+F N W+ FA+YYSL +G
Sbjct: 274 RIPENFITMFGNELENVATVTVPDGCDWEMDLKKCGEDVYFCNKEWQQFAEYYSLRYGCF 453
Query: 68 VLFEYKDTSHFGVHIFDKSTLEIRYPSHENQDEEHNPDQIDDDSVEILDKIPPCKKTKLK 127
+ F Y+ S+F V IFD +++EI YP K P T +
Sbjct: 454 LSFRYEGNSNFSVIIFDATSVEICYPL----------------------KTP---STSGE 558
Query: 128 SPMSCPKPRKKLRTSTSEDVGE 149
+ CP+P K+ + TSE G+
Sbjct: 559 TNTECPRPMKRSKVETSESPGK 624
>TC79718
Length = 1377
Score = 80.1 bits (196), Expect = 2e-15
Identities = 69/293 (23%), Positives = 119/293 (40%), Gaps = 30/293 (10%)
Frame = +2
Query: 10 IPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNGWKDFAKYYSLDHGHMVL 69
+P F+ + LK P G W I + D V+F +GW+ F +SL ++
Sbjct: 191 LPKTFSDNLKKKLPENVTLKGPSGVVWNIGLTTRDNTVYFVDGWQQFVNDHSLKENDFLV 370
Query: 70 FEYKDTSHFGVHIFDKSTLEIRYPSH-----ENQDEEHNPDQIDDDSVEILDKIPPCKKT 124
F+Y S F V IFD ++ + S+ + E + D++ +
Sbjct: 371 FKYNGESLFEVLIFDGNSFCEKATSYFVGKCGHAQTEQGDSKAKDNNTSAFNAGVESASP 550
Query: 125 KLKSPMSCPKPRKKLRTSTSEDVGESPELHNLPKQVK------IEDDAG----RTTECLE 174
++ ++ P +TS+ + P + P Q K DD+G R
Sbjct: 551 QVADVVAKTTPAAVPSQTTSKRTKKKPVIEVTPVQTKKRGRPPKSDDSGEKLLRDLVACN 730
Query: 175 GEDLTSKITEALNR------ARTFQSKFPSFMTVMKPAYVNG-YSLHIPSFFAKKYLKN- 226
E + + + + A +F S FP F+ ++K V G +L IP F+ +L +
Sbjct: 731 KEHSEASTLDRIRKEDEKKIAESFTSSFPYFVKILKAGNVGGSRTLRIPYHFSAAHLPDF 910
Query: 227 ETDVLLQVLDGRTWSVSF-------NLGKFNAGWKKFTSVNNLKVGDVCLFEL 272
+ +V L+ G+ W+V+ + F GW F N + GD C+FEL
Sbjct: 911 KIEVTLRNSKGKCWTVNSVPCAKGKIIHSFCGGWMAFVRDNGVNFGDTCIFEL 1069
Score = 35.8 bits (81), Expect = 0.035
Identities = 29/109 (26%), Positives = 47/109 (42%), Gaps = 3/109 (2%)
Frame = +2
Query: 349 ANKFNSNNPFFIVNIAPDKNRDYRP-RVPKLFIRKHFNDIQQSEQTVILQCEKKLCPV-- 405
A F S+ P+F+ + R R+P F H D + V L+ K C
Sbjct: 791 AESFTSSFPYFVKILKAGNVGGSRTLRIPYHFSAAHLPDFKIE---VTLRNSKGKCWTVN 961
Query: 406 KLTCYPKKGPSRLSKGWVEFAKKSKLEVGDACVFELINKEDPVLDVHIY 454
+ C K GW+ F + + + GD C+FEL+ + V+ V+I+
Sbjct: 962 SVPCAKGKIIHSFCGGWMAFVRDNGVNFGDTCIFELVT--NYVMQVYIF 1102
>TC79503 weakly similar to GP|21734794|gb|AAM76972.1 reduced vernalization
response 1 {Arabidopsis thaliana}, partial (6%)
Length = 910
Score = 73.2 bits (178), Expect = 2e-13
Identities = 34/92 (36%), Positives = 55/92 (58%)
Frame = +3
Query: 3 MESFVQRIPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNGWKDFAKYYSL 62
++S RIP+NF +++G ++ T L PDGT W + K D + F +GW+DF + YS+
Sbjct: 147 LQSKQLRIPDNFLRKHGAQLSTITTLTVPDGTVWHLRLKKVDNQICFVDGWQDFVQRYSI 326
Query: 63 DHGHMVLFEYKDTSHFGVHIFDKSTLEIRYPS 94
G ++F + +F VH+F+ ST E+ Y S
Sbjct: 327 GIG*FLVFTCEGNLNFIVHMFNISTAELNYQS 422
>TC79504 weakly similar to GP|21734794|gb|AAM76972.1 reduced vernalization
response 1 {Arabidopsis thaliana}, partial (26%)
Length = 695
Score = 70.1 bits (170), Expect = 2e-12
Identities = 40/126 (31%), Positives = 68/126 (53%), Gaps = 9/126 (7%)
Frame = +1
Query: 3 MESFVQRIPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNGWKDFAKYYSL 62
++S RIP+NF +++G ++ L P G+ W++ K D + F +GW+DF + YS+
Sbjct: 133 LQSKQLRIPDNFLRKHGDQLSTVATLTIPGGSVWRLGLKKVDNSICFVDGWQDFVQRYSI 312
Query: 63 DHGHMVLFEYKDTSHFGVHIFDKSTLEIRYPSHENQDEE--------HNPDQIDD-DSVE 113
G++++F Y+ S+F +HIF T E+ Y S E + + I+D DS +
Sbjct: 313 GIGYLLVFIYEGRSNFIIHIF--RTAELNYDSTMKSRTEGPFYEAYNYVFEGIEDIDSFD 486
Query: 114 ILDKIP 119
LD P
Sbjct: 487 FLDSAP 504
>TC80302
Length = 886
Score = 56.6 bits (135), Expect = 2e-08
Identities = 47/184 (25%), Positives = 82/184 (44%), Gaps = 9/184 (4%)
Frame = +3
Query: 134 KPRKKLRTSTSEDVGESPELHNLPKQVKIEDDAGRTTECLEGEDLTSKITEALNRARTFQ 193
KP ++ ++ G P+ N ++ ++ A + S + A++F
Sbjct: 114 KPANEVTPGQTKKRGRPPKEGN-SREGALDLRASNKEHSEAAQSRLSSAKDEKKLAQSFT 290
Query: 194 SKFPSFMTVMKPAYVNGYS-LHIPSFFAKKYLKN-ETDVLLQVLDGRTWSVS-------F 244
S FP F+ ++K V+G L++P F+ +L N + ++L+ L G W+ +
Sbjct: 291 STFPYFVKIIKTFNVDGPRILNVPHQFSIAHLPNGKIKIILRNLKGEQWTANSVPRSRVH 470
Query: 245 NLGKFNAGWKKFTSVNNLKVGDVCLFELNKSVPLSLKVLIFPLAEEPHSLPRSLVQGDRV 304
GW F NN+K+GDVC+FEL L ++V AE S VQ + +
Sbjct: 471 TSHTLCGGWMSFVRANNIKLGDVCVFELINDCELRVRV-----AEVDEDGLVSEVQKEGI 635
Query: 305 NHIN 308
+H N
Sbjct: 636 DHQN 647
Score = 35.8 bits (81), Expect = 0.035
Identities = 13/26 (50%), Positives = 19/26 (73%)
Frame = +3
Query: 418 LSKGWVEFAKKSKLEVGDACVFELIN 443
L GW+ F + + +++GD CVFELIN
Sbjct: 483 LCGGWMSFVRANNIKLGDVCVFELIN 560
>CA921493
Length = 656
Score = 51.2 bits (121), Expect = 8e-07
Identities = 32/93 (34%), Positives = 48/93 (51%), Gaps = 9/93 (9%)
Frame = -1
Query: 189 ARTFQSKFPSFMTVMKPAYVNGY-SLHIPSFFAKKYLKNE-TDVLLQVLDGRTWSVSF-- 244
A++F S FP F+ ++K V G +L IP F+ +L ++ T+V L+ G W+V+
Sbjct: 587 AQSFTSSFPYFVKILKNGNVGGSRTLRIPRRFSAAHLPDDKTEVTLRNSRGECWTVNSVP 408
Query: 245 -----NLGKFNAGWKKFTSVNNLKVGDVCLFEL 272
L F GW F N + GD C+FEL
Sbjct: 407 YAKRGMLHTFCGGWMSFVRDNGVNFGDTCIFEL 309
Score = 45.1 bits (105), Expect = 6e-05
Identities = 33/109 (30%), Positives = 51/109 (46%), Gaps = 3/109 (2%)
Frame = -1
Query: 349 ANKFNSNNPFFIVNIAPDKNRDYRP-RVPKLFIRKHFNDIQQSEQTVILQCEKKLC-PVK 406
A F S+ P+F+ + R R+P+ F H D + V L+ + C V
Sbjct: 587 AQSFTSSFPYFVKILKNGNVGGSRTLRIPRRFSAAHLPD---DKTEVTLRNSRGECWTVN 417
Query: 407 LTCYPKKGPSR-LSKGWVEFAKKSKLEVGDACVFELINKEDPVLDVHIY 454
Y K+G GW+ F + + + GD C+FEL++ D V+ VHIY
Sbjct: 416 SVPYAKRGMLHTFCGGWMSFVRDNGVNFGDTCIFELVS--DYVMQVHIY 276
>TC89357 similar to GP|20145851|emb|CAD29616. auxin response factor 36
{Arabidopsis thaliana}, partial (21%)
Length = 843
Score = 51.2 bits (121), Expect = 8e-07
Identities = 25/75 (33%), Positives = 34/75 (45%)
Frame = +3
Query: 10 IPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNGWKDFAKYYSLDHGHMVL 69
+P F+ + LK P G W I + DG +F GW+ F K +SL +
Sbjct: 267 LPKTFSNNVKKKLPENVVLKGPGGVTWNIGLTSRDGTFYFTQGWEQFVKDHSLKENDFLF 446
Query: 70 FEYKDTSHFGVHIFD 84
F+Y S F V IFD
Sbjct: 447 FKYNGESLFEVLIFD 491
>TC81996 similar to GP|20145851|emb|CAD29616. auxin response factor 36
{Arabidopsis thaliana}, partial (15%)
Length = 700
Score = 49.7 bits (117), Expect = 2e-06
Identities = 25/76 (32%), Positives = 37/76 (47%)
Frame = +3
Query: 10 IPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNGWKDFAKYYSLDHGHMVL 69
+P F+ + LK P G W I + D V+F +GW+ F K +SL ++
Sbjct: 147 LPKTFSDNLKNKLPENVTLKGPSGVVWNIGLTTRDYTVYFTDGWQRFVKDHSLKENDFLV 326
Query: 70 FEYKDTSHFGVHIFDK 85
F+Y S F V IFD+
Sbjct: 327 FKYNGESLFEVLIFDR 374
>TC81461
Length = 745
Score = 49.3 bits (116), Expect = 3e-06
Identities = 25/75 (33%), Positives = 34/75 (45%)
Frame = +1
Query: 10 IPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNGWKDFAKYYSLDHGHMVL 69
+P F+ + LK P G W I + D V+F NGW+ F +SL +
Sbjct: 238 LPKTFSDNLKKKLPENVTLKGPSGVVWNIGLTTIDDTVYFTNGWQQFVNDHSLKESDFLF 417
Query: 70 FEYKDTSHFGVHIFD 84
F+Y S F V IFD
Sbjct: 418 FKYNGESLFEVLIFD 462
>AW573956 similar to GP|20145851|emb auxin response factor 36 {Arabidopsis
thaliana}, partial (15%)
Length = 555
Score = 48.1 bits (113), Expect = 7e-06
Identities = 25/75 (33%), Positives = 35/75 (46%)
Frame = +2
Query: 10 IPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNGWKDFAKYYSLDHGHMVL 69
+P F+ + LK P G W I + D V+F GW+ F K +SL ++
Sbjct: 209 LPKTFSDNLKKKLPENVTLKGPSGAVWDIGLTTRDNTVYFVGGWERFVKDHSLKENDFLV 388
Query: 70 FEYKDTSHFGVHIFD 84
F+Y S F V IFD
Sbjct: 389 FKYNGESLFEVLIFD 433
>BG646843 similar to GP|20145851|emb auxin response factor 36 {Arabidopsis
thaliana}, partial (5%)
Length = 758
Score = 47.8 bits (112), Expect = 9e-06
Identities = 23/74 (31%), Positives = 35/74 (47%)
Frame = +3
Query: 10 IPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNGWKDFAKYYSLDHGHMVL 69
+P F+ + LK P G W + + D ++F NGW+ F K +SL ++
Sbjct: 231 LPKTFSDNLKKKLPGNVTLKGPSGVVWNVGLTARDDTIYFTNGWQRFVKDHSLKENDFLV 410
Query: 70 FEYKDTSHFGVHIF 83
F+Y S F V IF
Sbjct: 411 FKYNGESLFEVLIF 452
>TC77100
Length = 926
Score = 42.0 bits (97), Expect = 5e-04
Identities = 33/115 (28%), Positives = 55/115 (47%), Gaps = 8/115 (6%)
Frame = +1
Query: 197 PSFMTVMKPAYVNG-YSLHIPSFFAKKYLKNETDVLLQVLDGRTWSVSFN----LGKFNA 251
P F V+ +++ Y + S ++ E +L+ G++W +++N +F++
Sbjct: 280 PYFHVVLSKTHLSTRYGMGPSSSICEELPSKEVPTILKYR-GKSWGMTYNGQNKTKQFDS 456
Query: 252 -GWKKFTSVNNLKVGDVCLFEL--NKSVPLSLKVLIFPLAEEPHSLPRSLVQGDR 303
W+KF N LK+GD C+FEL N + KV I EEP L G+R
Sbjct: 457 VSWEKFAEDNYLKLGDACVFELMKNSEEEIVFKVQILRGEEEPILLSEFPGTGER 621
Score = 37.7 bits (86), Expect = 0.009
Identities = 19/40 (47%), Positives = 28/40 (69%), Gaps = 2/40 (5%)
Frame = +1
Query: 419 SKGWVEFAKKSKLEVGDACVFELI--NKEDPVLDVHIYRG 456
S W +FA+ + L++GDACVFEL+ ++E+ V V I RG
Sbjct: 454 SVSWEKFAEDNYLKLGDACVFELMKNSEEEIVFKVQILRG 573
>CA990321 weakly similar to PIR|S03170|S03 homeotic protein cut - fruit fly
(Drosophila melanogaster), partial (2%)
Length = 451
Score = 42.0 bits (97), Expect = 5e-04
Identities = 28/110 (25%), Positives = 47/110 (42%), Gaps = 8/110 (7%)
Frame = +3
Query: 9 RIPNNFTK--RYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNGWKDFAKYYSLDHGH 66
RIPN F R + L+ G DW + +++F GWK F + SL+
Sbjct: 69 RIPNAFVNLMRSKRKVIKDFILRDRRGRDWHVKARLIGDELYFDGGWKQFREENSLEEDD 248
Query: 67 MVLFEYKDTSHFGVHIF------DKSTLEIRYPSHENQDEEHNPDQIDDD 110
++F + + + F I +K + + ++ +DEE D DDD
Sbjct: 249 FLVFTHIENTVFRFKILELSSMCEKKKVSVVEENNNMEDEEEVNDGDDDD 398
>TC92612
Length = 457
Score = 41.2 bits (95), Expect = 8e-04
Identities = 26/86 (30%), Positives = 44/86 (50%), Gaps = 9/86 (10%)
Frame = +3
Query: 189 ARTFQSKFPSFMTVMKPAYVNG-YSLHIPSFFAKKYLKN-ETDVLLQVLDGRTWSVS--- 243
A++F S FP F+ ++K V+G Y+L++P F+ + N +T ++L L G W+ +
Sbjct: 198 AQSFTSSFPYFVKIIKTFNVSGSYTLNVPYQFSMSHFPNCKTKIILYNLKGEHWTGNSVP 377
Query: 244 ----FNLGKFNAGWKKFTSVNNLKVG 265
GW F N++KVG
Sbjct: 378 TTRVHTSHTLCGGWMAFVRGNSIKVG 455
>TC76590 similar to GP|14532680|gb|AAK64141.1 unknown protein {Arabidopsis
thaliana}, partial (4%)
Length = 1521
Score = 39.3 bits (90), Expect = 0.003
Identities = 28/103 (27%), Positives = 48/103 (46%), Gaps = 6/103 (5%)
Frame = +1
Query: 189 ARTFQSKFPSFMTVMKPAYVNGYSLHIPSFFAKKYLKNETDVLLQVLDGRTWSVSFNLGK 248
A +++ P F++ + G + I S FA ++L +LL+ DG+ W S + +
Sbjct: 997 AEAYRTANPHFISKV----TRGNNAFIDSTFASRHLNENVSILLRNSDGQEWEDSAEMFR 1164
Query: 249 FNAGWKKFTSV------NNLKVGDVCLFELNKSVPLSLKVLIF 285
++ KF N L D C FEL + P+ L V++F
Sbjct: 1165 KDSHQMKFKKFHIFKNDNYLCQEDYCAFELIQINPVVLNVIMF 1293
>TC76598 similar to GP|14532680|gb|AAK64141.1 unknown protein {Arabidopsis
thaliana}, partial (4%)
Length = 1222
Score = 38.5 bits (88), Expect = 0.005
Identities = 28/103 (27%), Positives = 47/103 (45%), Gaps = 6/103 (5%)
Frame = +3
Query: 189 ARTFQSKFPSFMTVMKPAYVNGYSLHIPSFFAKKYLKNETDVLLQVLDGRTWSVSFNLGK 248
A +++ P F++ + G + I S FA ++L +LL+ DG+ W S + +
Sbjct: 924 AEAYRTANPHFISKV----TRGNNAFIDSTFASRHLNENVSILLRNSDGQEWEDSAEMFR 1091
Query: 249 FNAGWKKFTSV------NNLKVGDVCLFELNKSVPLSLKVLIF 285
++ KF N L D C FEL P+ L V++F
Sbjct: 1092 KDSHQMKFKKFHIFKNDNYLCQEDYCAFELIXINPVVLNVIMF 1220
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.136 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,648,084
Number of Sequences: 36976
Number of extensions: 238726
Number of successful extensions: 1263
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 1209
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1252
length of query: 457
length of database: 9,014,727
effective HSP length: 99
effective length of query: 358
effective length of database: 5,354,103
effective search space: 1916768874
effective search space used: 1916768874
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0040.4


