

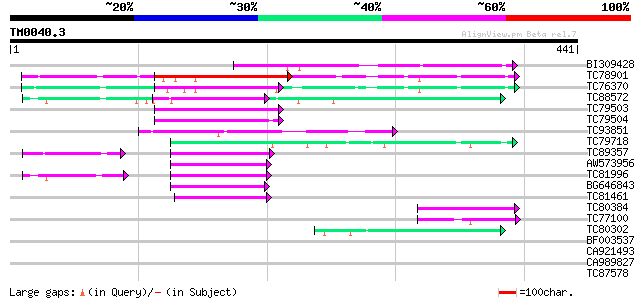
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0040.3
(441 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BI309428 weakly similar to GP|21734794|gb| reduced vernalization... 112 4e-25
TC78901 similar to GP|21734794|gb|AAM76972.1 reduced vernalizati... 98 7e-21
TC76370 similar to GP|21734794|gb|AAM76972.1 reduced vernalizati... 88 6e-18
TC88572 weakly similar to GP|20145851|emb|CAD29616. auxin respon... 80 1e-15
TC79503 weakly similar to GP|21734794|gb|AAM76972.1 reduced vern... 72 3e-13
TC79504 weakly similar to GP|21734794|gb|AAM76972.1 reduced vern... 69 4e-12
TC93851 weakly similar to GP|21734794|gb|AAM76972.1 reduced vern... 68 8e-12
TC79718 65 7e-11
TC89357 similar to GP|20145851|emb|CAD29616. auxin response fact... 56 2e-08
AW573956 similar to GP|20145851|emb auxin response factor 36 {Ar... 50 1e-06
TC81996 similar to GP|20145851|emb|CAD29616. auxin response fact... 50 2e-06
BG646843 similar to GP|20145851|emb auxin response factor 36 {Ar... 48 7e-06
TC81461 46 3e-05
TC80384 44 1e-04
TC77100 42 4e-04
TC80302 42 6e-04
BF003537 40 0.001
CA921493 40 0.002
CA989827 37 0.020
TC87578 similar to PIR|T12113|T12113 transcription factor - fava... 36 0.026
>BI309428 weakly similar to GP|21734794|gb| reduced vernalization response 1
{Arabidopsis thaliana}, partial (6%)
Length = 710
Score = 112 bits (279), Expect = 4e-25
Identities = 78/231 (33%), Positives = 119/231 (50%), Gaps = 10/231 (4%)
Frame = +1
Query: 175 FVKYYSLDHGHLVLFEFKDASHFGVHIFDNSTLEIEYPSN-----ENQGEEHNPD----Q 225
F + YSL HG LV+FE+K F V I + LEI+Y ++ EN +H+ D +
Sbjct: 4 FTENYSLGHGCLVVFEYKGTFKFDVVILGKNALEIDYDTSCDSDDENDNVDHSDDDESVE 183
Query: 226 IGDEILLPKKTKPKSPMSLSRPCKKLRTGKSKDVGKSPELRKRKKVQVEGGEPSSLGPHF 285
I DE L KK + +SP SRP KK++ K ++ + K V+
Sbjct: 184 ILDEWLNRKKARQRSPFVSSRPHKKVQGDDEKTTKRTSSMNWPKDVR------------- 324
Query: 286 TAQGEARKYTPKNPSFTVSLKPVIKLAYRPPVPISFIREYLNEKKQTIKLKIEENLWPVE 345
AQ A+ + NP FT+ +KP+ + + VP ++ + K + L I + W V+
Sbjct: 325 -AQEVAQNFISCNPFFTILIKPINLVEHTRSVPD--LKGVIENKDTNVMLLIGKRSWNVK 495
Query: 346 FIYYPEHGSGK-LSKGWIQFAKESKLVGGDVCVFELINKDEENPVLEVHIF 395
+ E +G+ LS GW FA+ES L GDVCVFELINK ++ V ++H++
Sbjct: 496 LLRSYEGKNGRRLSAGWYLFARESGLKSGDVCVFELINK--KDLVFKIHVY 642
Score = 37.0 bits (84), Expect = 0.015
Identities = 17/26 (65%), Positives = 20/26 (76%)
Frame = +1
Query: 73 LVLFEFKGPSKFEVCIFDKNALEIKY 98
LV+FE+KG KF+V I KNALEI Y
Sbjct: 37 LVVFEYKGTFKFDVVILGKNALEIDY 114
>TC78901 similar to GP|21734794|gb|AAM76972.1 reduced vernalization response
1 {Arabidopsis thaliana}, partial (37%)
Length = 1760
Score = 97.8 bits (242), Expect = 7e-21
Identities = 45/108 (41%), Positives = 66/108 (60%)
Frame = +2
Query: 113 FFRIITPPCLEAAELILPNGFMKKYSGGMSNPVFLKAPDNTSWEIHWSKHDSYFWFQKGW 172
F +++ P L+A +L LP+ FM+KY G +S V L PD + W + K D+ FWF GW
Sbjct: 221 FHKLVLPSTLQAKQLRLPDDFMRKYGGDISPTVTLTVPDGSVWRVIMKKVDNKFWFLDGW 400
Query: 173 KEFVKYYSLDHGHLVLFEFKDASHFGVHIFDNSTLEIEYPSNENQGEE 220
EFV+ YS+ G+L++F+++ SHF V+IF T EI Y S + E
Sbjct: 401 NEFVQNYSISTGYLLVFKYEGKSHFTVNIFSLPTSEINYQSPAQRSNE 544
Score = 94.4 bits (233), Expect = 8e-20
Identities = 109/405 (26%), Positives = 171/405 (41%), Gaps = 18/405 (4%)
Frame = +2
Query: 10 KLPNAFTRKYSGDNMSNPVFLKPLDNISWEINWSKQDSDIWLQNVWEEFVKLYYLDQGHM 69
+LP+ F RKY GD +S V L D W + K D+ W + W EFV+ Y + G+
Sbjct: 266 RLPDDFMRKYGGD-ISPTVTLTVPDGSVWRVIMKKVDNKFWFLDGWNEFVQNYSISTGY- 439
Query: 70 VWHLVLFEFKGPSKFEVCIFDKNALEIKYLPLSNKQIPNSATRFFRIIT-------PPCL 122
L++F+++G S F V IF EI Y S Q N A+ F + +T +
Sbjct: 440 ---LLVFKYEGKSHFTVNIFSLPTSEINYQ--SPAQRSNEASLFGKRLTIFEEMEDEDSV 604
Query: 123 EAAEL----ILPNGFMKKYSGGMSN---PVFLKAPDNTSWEIHWSKHDSYFWFQKGWKEF 175
E E + P+ K G ++ P + P + SK +S W + G
Sbjct: 605 EIMESSPTKLTPSLLQNKAVSGSADKLTPGKSRPPPALQNLFNGSKLNSINWGEGGNTPS 784
Query: 176 VKYYSLDHGHLVLFEFKDASHFGVHIFDNSTLEIEYPSNENQGEEHNPDQIGDEILLPKK 235
S+D+ + G+ + +E + + E + +++ + +K
Sbjct: 785 RNDNSVDN--------QLTRDIGLQF---NVVEFKKSNEELKLRAATDEKVKKTAVKKRK 931
Query: 236 TKPKSPMSLSRPCKKLRTGKSKDVGKSPELRKRKKVQVEGGEPSSLGPHFTAQGEARKYT 295
+ + P S ++R +S RKR E + A A+ +
Sbjct: 932 SDVQEPPSEHEDEVEMRNR----FYESASARKRTATAEEREK---------AINAAKTFE 1072
Query: 296 PKNPSFTVSLKPVIKLAYRPPV---PISFIREYLNEKKQTIKLKIEENL-WPVEFIYYPE 351
P NP V L+P YR + P F + LN IKL+I + WPV +Y
Sbjct: 1073PSNPFCRVVLRP--SYLYRGCIMYLPSCFAEKNLNGVSGIIKLQISDGRQWPVRCLY--R 1240
Query: 352 HGSGKLSKGWIQFAKESKLVGGDVCVFELINKDEENPVLEVHIFK 396
G KLS+GW +F+ E+ L GDVCVFEL+ E VL+V +F+
Sbjct: 1241GGRAKLSQGWFEFSLENNLGEGDVCVFELVATKE--VVLQVTVFR 1369
>TC76370 similar to GP|21734794|gb|AAM76972.1 reduced vernalization response
1 {Arabidopsis thaliana}, partial (36%)
Length = 1840
Score = 88.2 bits (217), Expect = 6e-18
Identities = 103/405 (25%), Positives = 162/405 (39%), Gaps = 18/405 (4%)
Frame = +1
Query: 10 KLPNAFTRKYSGDNMSNPVFLKPLDNISWEINWSKQDSDIWLQNVWEEFVKLYYLDQGHM 69
++P+ F RKY G +S L D W + K D+ IW + W++FV+ Y + G+
Sbjct: 190 RIPDNFLRKY-GAQLSTIATLTVPDGTVWRLGLKKVDNRIWFVDGWQDFVQRYAIGIGYF 366
Query: 70 VWHLVLFEFKGPSKFEVCIFDKNALEIKYLPLSNKQIPNSATRFFRIITPPC-------- 121
+ +F ++G S F V IF+ + E+ Y + + I
Sbjct: 367 L----VFTYEGNSNFIVHIFNMSTAELNYQSAMRSRTEGPCYANYHPIFEDIEDIDSFEF 534
Query: 122 LEAAELILPNGFM--KKYSGGMSN--PVFLKAPDNTSWEIHWSKHDSYFWFQKGWKEFVK 177
L+++ L G + K +SG + P P + SK + W G
Sbjct: 535 LDSSPSNLTPGALQDKVFSGSLDQLTPAKNHTPSLQNLFNGGSKLNHVNWGDIGG----- 699
Query: 178 YYSLDHGHLVLFEFKDASHFGVHIFDNST--LEIEYPSNENQGEEHNPDQIGDEILLPKK 235
+L + F + F ST L++ YP+ E D +K
Sbjct: 700 --TLSSKSAIQSTRDIGVQFNANEFKKSTEGLKLRYPNEEGVNNT-------DTKKTSRK 852
Query: 236 TKPKSPMSLSRPCKKLRTGKSKDVGKSPELRKRKKVQVEGGEPSSLGPHFTAQGEARKYT 295
+ P + + + +S RKR V E E A E++ +
Sbjct: 853 KRKSDPSAQEATAENDEEAERYRFYESASARKRT-VTAEERE--------RAINESKTFE 1005
Query: 296 PKNPSFTVSLKPVIKLAYRPPV---PISFIREYLNEKKQTIKLKIEENL-WPVEFIYYPE 351
P NP V L+P YR + P F + LN IKL+I + WPV +Y +
Sbjct: 1006PTNPFCRVVLRP--SYLYRGCIMYLPSCFAEKNLNGVSGFIKLQISDGRQWPVRCLY--K 1173
Query: 352 HGSGKLSKGWIQFAKESKLVGGDVCVFELINKDEENPVLEVHIFK 396
G KLS+GW +F E+ L GD+CVFEL+ E VL+V +F+
Sbjct: 1174GGRAKLSQGWYEFTLENNLGEGDICVFELLRTRE--VVLQVTLFR 1302
Score = 85.1 bits (209), Expect = 5e-17
Identities = 37/101 (36%), Positives = 61/101 (59%)
Frame = +1
Query: 113 FFRIITPPCLEAAELILPNGFMKKYSGGMSNPVFLKAPDNTSWEIHWSKHDSYFWFQKGW 172
F ++I P L+ +L +P+ F++KY +S L PD T W + K D+ WF GW
Sbjct: 145 FHKLILPSSLQTRQLRIPDNFLRKYGAQLSTIATLTVPDGTVWRLGLKKVDNRIWFVDGW 324
Query: 173 KEFVKYYSLDHGHLVLFEFKDASHFGVHIFDNSTLEIEYPS 213
++FV+ Y++ G+ ++F ++ S+F VHIF+ ST E+ Y S
Sbjct: 325 QDFVQRYAIGIGYFLVFTYEGNSNFIVHIFNMSTAELNYQS 447
>TC88572 weakly similar to GP|20145851|emb|CAD29616. auxin response factor
36 {Arabidopsis thaliana}, partial (21%)
Length = 1755
Score = 80.5 bits (197), Expect = 1e-15
Identities = 107/431 (24%), Positives = 165/431 (37%), Gaps = 56/431 (12%)
Frame = +2
Query: 11 LPNAFTRKYSGDNMSNP----VFLKPLDNISWEINWSKQDSDIWLQNVWEEFVKLYYLDQ 66
LP F+ DN+ N V LK + W+I + +D I+ + W++FVK + L Q
Sbjct: 137 LPKTFS-----DNLKNKLPANVTLKGPSGVVWDIGLTTRDDTIYSTDGWQQFVKDHSLKQ 301
Query: 67 GHMVWHLVLFEFKGPSKFEVCIFDKNALEIK---YLPLSNKQ------------------ 105
+ +F++ G S FEV IF + K Y L Q
Sbjct: 302 NDFL----VFKYNGESLFEVLIFHGESFCEKAASYFILERGQAHTGQRGRKAKDTNTSVE 469
Query: 106 -------------IPNSA-------TRFFRIITPPCLEA---AELILPNGFMKKYSGGMS 142
+P + T F I L A +L LP F +
Sbjct: 470 EVNTPSNGSVECGLPEKSWEEDIYWTHFQFIHFTQFLRADFEQQLALPKTFSDNLKKKLP 649
Query: 143 NPVFLKAPDNTSWEIHWSKHDSYFWFQKGWKEFVKYYSLDHGHLVLFEFKDASHFGVHIF 202
V LK P W I + D +F GWK F+K +SL ++F++ S F V IF
Sbjct: 650 ENVTLKGPSGVVWNIGLTTRDDIVYFVDGWKRFIKDHSLKQNDFLVFKYNGKSLFEVLIF 829
Query: 203 DNSTLEIEYPSNENQGEEHNP--DQIGDEIL-LPKKTKPKSPMSLSRPCKK--LRTGKSK 257
D + E ++ G+ N +Q G + K P+ ++ + P + RT K
Sbjct: 830 DGDSF-CEKAASYFVGKCGNAQIEQGGRKAKDTNKSVSPEQFLADAVPSQTNGKRTKKRP 1006
Query: 258 DVGKSPELRKRKKVQVEGGEPSSLGPHFTAQGEARKYTPKNPSFTVSLKPVIKLAYRP-P 316
+P KR + S L + A +T P F +K R
Sbjct: 1007VNEVTPLQTKRGRRPKAEAALSKLSSSHAEKKIAESFTSSFPYFVKMIKTFNVDGPRILN 1186
Query: 317 VPISFIREYL-NEKKQTIKLKIEENLWPVEFIYYPEHGSGK-LSKGWIQFAKESKLVGGD 374
VP F + +L N K + I ++ W V + + L GW+ F + + + GD
Sbjct: 1187VPHQFSKAHLPNRKIKIILHNLKGEQWTVNSVPTTRVNTDHTLCGGWVNFVRGNNIKVGD 1366
Query: 375 VCVFELINKDE 385
VC+FELI++ E
Sbjct: 1367VCIFELIHECE 1399
Score = 50.4 bits (119), Expect = 1e-06
Identities = 29/91 (31%), Positives = 43/91 (46%)
Frame = +2
Query: 112 RFFRIITPPCLEAAELILPNGFMKKYSGGMSNPVFLKAPDNTSWEIHWSKHDSYFWFQKG 171
RF + + P LE +L LP F + V LK P W+I + D + G
Sbjct: 89 RFTQFLQPTHLEQ-QLALPKTFSDNLKNKLPANVTLKGPSGVVWDIGLTTRDDTIYSTDG 265
Query: 172 WKEFVKYYSLDHGHLVLFEFKDASHFGVHIF 202
W++FVK +SL ++F++ S F V IF
Sbjct: 266 WQQFVKDHSLKQNDFLVFKYNGESLFEVLIF 358
>TC79503 weakly similar to GP|21734794|gb|AAM76972.1 reduced vernalization
response 1 {Arabidopsis thaliana}, partial (6%)
Length = 910
Score = 72.4 bits (176), Expect = 3e-13
Identities = 34/101 (33%), Positives = 58/101 (56%)
Frame = +3
Query: 113 FFRIITPPCLEAAELILPNGFMKKYSGGMSNPVFLKAPDNTSWEIHWSKHDSYFWFQKGW 172
F ++I P L++ +L +P+ F++K+ +S L PD T W + K D+ F GW
Sbjct: 120 FHKLIFPTTLQSKQLRIPDNFLRKHGAQLSTITTLTVPDGTVWHLRLKKVDNQICFVDGW 299
Query: 173 KEFVKYYSLDHGHLVLFEFKDASHFGVHIFDNSTLEIEYPS 213
++FV+ YS+ G ++F + +F VH+F+ ST E+ Y S
Sbjct: 300 QDFVQRYSIGIG*FLVFTCEGNLNFIVHMFNISTAELNYQS 422
Score = 37.4 bits (85), Expect = 0.012
Identities = 24/89 (26%), Positives = 43/89 (47%)
Frame = +3
Query: 10 KLPNAFTRKYSGDNMSNPVFLKPLDNISWEINWSKQDSDIWLQNVWEEFVKLYYLDQGHM 69
++P+ F RK+ G +S L D W + K D+ I + W++FV+ Y + G
Sbjct: 165 RIPDNFLRKH-GAQLSTITTLTVPDGTVWHLRLKKVDNQICFVDGWQDFVQRYSIGIG-- 335
Query: 70 VWHLVLFEFKGPSKFEVCIFDKNALEIKY 98
++F +G F V +F+ + E+ Y
Sbjct: 336 --*FLVFTCEGNLNFIVHMFNISTAELNY 416
>TC79504 weakly similar to GP|21734794|gb|AAM76972.1 reduced vernalization
response 1 {Arabidopsis thaliana}, partial (26%)
Length = 695
Score = 68.9 bits (167), Expect = 4e-12
Identities = 33/101 (32%), Positives = 59/101 (57%)
Frame = +1
Query: 113 FFRIITPPCLEAAELILPNGFMKKYSGGMSNPVFLKAPDNTSWEIHWSKHDSYFWFQKGW 172
F ++I P L++ +L +P+ F++K+ +S L P + W + K D+ F GW
Sbjct: 106 FHKLIFPTTLQSKQLRIPDNFLRKHGDQLSTVATLTIPGGSVWRLGLKKVDNSICFVDGW 285
Query: 173 KEFVKYYSLDHGHLVLFEFKDASHFGVHIFDNSTLEIEYPS 213
++FV+ YS+ G+L++F ++ S+F +HIF T E+ Y S
Sbjct: 286 QDFVQRYSIGIGYLLVFIYEGRSNFIIHIF--RTAELNYDS 402
Score = 40.8 bits (94), Expect = 0.001
Identities = 24/80 (30%), Positives = 42/80 (52%)
Frame = +1
Query: 10 KLPNAFTRKYSGDNMSNPVFLKPLDNISWEINWSKQDSDIWLQNVWEEFVKLYYLDQGHM 69
++P+ F RK+ GD +S L W + K D+ I + W++FV+ Y + G
Sbjct: 151 RIPDNFLRKH-GDQLSTVATLTIPGGSVWRLGLKKVDNSICFVDGWQDFVQRYSIGIG-- 321
Query: 70 VWHLVLFEFKGPSKFEVCIF 89
+L++F ++G S F + IF
Sbjct: 322 --YLLVFIYEGRSNFIIHIF 375
>TC93851 weakly similar to GP|21734794|gb|AAM76972.1 reduced vernalization
response 1 {Arabidopsis thaliana}, partial (9%)
Length = 710
Score = 67.8 bits (164), Expect = 8e-12
Identities = 52/203 (25%), Positives = 85/203 (41%), Gaps = 2/203 (0%)
Frame = +1
Query: 101 LSNKQIPNSATRFFRIITPPCLEAAELILPNGFMKKYSGGMSNPVFLKAPDNTSWEIHWS 160
++ K++ S F + I P + E+ +P F+ + + N + PD WE+
Sbjct: 196 MAEKEVRESI-HFKKAILPSPIYDKEIRIPENFITMFGNELENVATVTVPDGCDWEMDLK 372
Query: 161 K--HDSYFWFQKGWKEFVKYYSLDHGHLVLFEFKDASHFGVHIFDNSTLEIEYPSNENQG 218
K D YF K W++F +YYSL +G + F ++ S+F V IFD +++EI YP
Sbjct: 373 KCGEDVYFC-NKEWQQFAEYYSLRYGCFLSFRYEGNSNFSVIIFDATSVEICYP------ 531
Query: 219 EEHNPDQIGDEILLPKKTKPKSPMSLSRPCKKLRTGKSKDVGKSPELRKRKKVQVEGGEP 278
L T ++ RP K+ + S+ GK +
Sbjct: 532 ------------LKTPSTSGETNTECPRPMKRSKVETSESPGKKV-------------KS 636
Query: 279 SSLGPHFTAQGEARKYTPKNPSF 301
S + A+ A + PKNP F
Sbjct: 637 MSNYAYKRAEDAANAFNPKNPHF 705
>TC79718
Length = 1377
Score = 64.7 bits (156), Expect = 7e-11
Identities = 79/316 (25%), Positives = 123/316 (38%), Gaps = 46/316 (14%)
Frame = +2
Query: 126 ELILPNGFMKKYSGGMSNPVFLKAPDNTSWEIHWSKHDSYFWFQKGWKEFVKYYSLDHGH 185
+L LP F + V LK P W I + D+ +F GW++FV +SL
Sbjct: 182 QLALPKTFSDNLKKKLPENVTLKGPSGVVWNIGLTTRDNTVYFVDGWQQFVNDHSLKEND 361
Query: 186 LVLFEFKDASHFGVHIFD------------------------NSTLEIEYPSNENQGEEH 221
++F++ S F V IFD +S + S N G E
Sbjct: 362 FLVFKYNGESLFEVLIFDGNSFCEKATSYFVGKCGHAQTEQGDSKAKDNNTSAFNAGVES 541
Query: 222 NPDQIGDEI-----------LLPKKTKPKSPMSLS--RPCKKLRTGKSKDVGKSPELRKR 268
Q+ D + K+TK K + ++ + K+ R KS D G+ LR
Sbjct: 542 ASPQVADVVAKTTPAAVPSQTTSKRTKKKPVIEVTPVQTKKRGRPPKSDDSGEK-LLRDL 718
Query: 269 KKVQVEGGEPSSLGPHFTAQGE---ARKYTPKNPSFTVSLKPVIKLAYRP-PVPISFIRE 324
E E S+L + E A +T P F LK R +P F
Sbjct: 719 VACNKEHSEASTL-DRIRKEDEKKIAESFTSSFPYFVKILKAGNVGGSRTLRIPYHFSAA 895
Query: 325 YLNEKKQTIKLKIEE-NLWPVEFIYYPEHGSGKL----SKGWIQFAKESKLVGGDVCVFE 379
+L + K + L+ + W V + GK+ GW+ F +++ + GD C+FE
Sbjct: 896 HLPDFKIEVTLRNSKGKCWTVNSV---PCAKGKIIHSFCGGWMAFVRDNGVNFGDTCIFE 1066
Query: 380 LINKDEENPVLEVHIF 395
L+ N V++V+IF
Sbjct: 1067LVT----NYVMQVYIF 1102
Score = 40.4 bits (93), Expect = 0.001
Identities = 24/83 (28%), Positives = 41/83 (48%)
Frame = +2
Query: 11 LPNAFTRKYSGDNMSNPVFLKPLDNISWEINWSKQDSDIWLQNVWEEFVKLYYLDQGHMV 70
LP F+ N V LK + W I + +D+ ++ + W++FV + L +
Sbjct: 191 LPKTFSDNLKKKLPEN-VTLKGPSGVVWNIGLTTRDNTVYFVDGWQQFVNDHSLKEN--- 358
Query: 71 WHLVLFEFKGPSKFEVCIFDKNA 93
++F++ G S FEV IFD N+
Sbjct: 359 -DFLVFKYNGESLFEVLIFDGNS 424
>TC89357 similar to GP|20145851|emb|CAD29616. auxin response factor 36
{Arabidopsis thaliana}, partial (21%)
Length = 843
Score = 56.2 bits (134), Expect = 2e-08
Identities = 27/81 (33%), Positives = 40/81 (49%)
Frame = +3
Query: 126 ELILPNGFMKKYSGGMSNPVFLKAPDNTSWEIHWSKHDSYFWFQKGWKEFVKYYSLDHGH 185
+L LP F + V LK P +W I + D F+F +GW++FVK +SL
Sbjct: 258 QLALPKTFSNNVKKKLPENVVLKGPGGVTWNIGLTSRDGTFYFTQGWEQFVKDHSLKEND 437
Query: 186 LVLFEFKDASHFGVHIFDNST 206
+ F++ S F V IFD +
Sbjct: 438 FLFFKYNGESLFEVLIFDGKS 500
Score = 42.4 bits (98), Expect = 4e-04
Identities = 25/80 (31%), Positives = 38/80 (47%)
Frame = +3
Query: 11 LPNAFTRKYSGDNMSNPVFLKPLDNISWEINWSKQDSDIWLQNVWEEFVKLYYLDQGHMV 70
LP F+ N V LK ++W I + +D + WE+FVK + L + +
Sbjct: 267 LPKTFSNNVKKKLPEN-VVLKGPGGVTWNIGLTSRDGTFYFTQGWEQFVKDHSLKENDFL 443
Query: 71 WHLVLFEFKGPSKFEVCIFD 90
+ F++ G S FEV IFD
Sbjct: 444 F----FKYNGESLFEVLIFD 491
>AW573956 similar to GP|20145851|emb auxin response factor 36 {Arabidopsis
thaliana}, partial (15%)
Length = 555
Score = 50.4 bits (119), Expect = 1e-06
Identities = 26/78 (33%), Positives = 38/78 (48%)
Frame = +2
Query: 126 ELILPNGFMKKYSGGMSNPVFLKAPDNTSWEIHWSKHDSYFWFQKGWKEFVKYYSLDHGH 185
+L LP F + V LK P W+I + D+ +F GW+ FVK +SL
Sbjct: 200 QLALPKTFSDNLKKKLPENVTLKGPSGAVWDIGLTTRDNTVYFVGGWERFVKDHSLKEND 379
Query: 186 LVLFEFKDASHFGVHIFD 203
++F++ S F V IFD
Sbjct: 380 FLVFKYNGESLFEVLIFD 433
Score = 39.7 bits (91), Expect = 0.002
Identities = 25/80 (31%), Positives = 38/80 (47%)
Frame = +2
Query: 11 LPNAFTRKYSGDNMSNPVFLKPLDNISWEINWSKQDSDIWLQNVWEEFVKLYYLDQGHMV 70
LP F+ N V LK W+I + +D+ ++ WE FVK + L +
Sbjct: 209 LPKTFSDNLKKKLPEN-VTLKGPSGAVWDIGLTTRDNTVYFVGGWERFVKDHSLKEN--- 376
Query: 71 WHLVLFEFKGPSKFEVCIFD 90
++F++ G S FEV IFD
Sbjct: 377 -DFLVFKYNGESLFEVLIFD 433
>TC81996 similar to GP|20145851|emb|CAD29616. auxin response factor 36
{Arabidopsis thaliana}, partial (15%)
Length = 700
Score = 50.1 bits (118), Expect = 2e-06
Identities = 26/78 (33%), Positives = 36/78 (45%)
Frame = +3
Query: 126 ELILPNGFMKKYSGGMSNPVFLKAPDNTSWEIHWSKHDSYFWFQKGWKEFVKYYSLDHGH 185
+L LP F + V LK P W I + D +F GW+ FVK +SL
Sbjct: 138 QLALPKTFSDNLKNKLPENVTLKGPSGVVWNIGLTTRDYTVYFTDGWQRFVKDHSLKEND 317
Query: 186 LVLFEFKDASHFGVHIFD 203
++F++ S F V IFD
Sbjct: 318 FLVFKYNGESLFEVLIFD 371
Score = 42.0 bits (97), Expect = 5e-04
Identities = 26/86 (30%), Positives = 43/86 (49%), Gaps = 4/86 (4%)
Frame = +3
Query: 11 LPNAFTRKYSGDNMSNP----VFLKPLDNISWEINWSKQDSDIWLQNVWEEFVKLYYLDQ 66
LP F+ DN+ N V LK + W I + +D ++ + W+ FVK + L +
Sbjct: 147 LPKTFS-----DNLKNKLPENVTLKGPSGVVWNIGLTTRDYTVYFTDGWQRFVKDHSLKE 311
Query: 67 GHMVWHLVLFEFKGPSKFEVCIFDKN 92
++F++ G S FEV IFD++
Sbjct: 312 N----DFLVFKYNGESLFEVLIFDRD 377
>BG646843 similar to GP|20145851|emb auxin response factor 36 {Arabidopsis
thaliana}, partial (5%)
Length = 758
Score = 48.1 bits (113), Expect = 7e-06
Identities = 24/77 (31%), Positives = 35/77 (45%)
Frame = +3
Query: 126 ELILPNGFMKKYSGGMSNPVFLKAPDNTSWEIHWSKHDSYFWFQKGWKEFVKYYSLDHGH 185
+L LP F + V LK P W + + D +F GW+ FVK +SL
Sbjct: 222 QLALPKTFSDNLKKKLPGNVTLKGPSGVVWNVGLTARDDTIYFTNGWQRFVKDHSLKEND 401
Query: 186 LVLFEFKDASHFGVHIF 202
++F++ S F V IF
Sbjct: 402 FLVFKYNGESLFEVLIF 452
Score = 39.3 bits (90), Expect = 0.003
Identities = 24/79 (30%), Positives = 37/79 (46%)
Frame = +3
Query: 11 LPNAFTRKYSGDNMSNPVFLKPLDNISWEINWSKQDSDIWLQNVWEEFVKLYYLDQGHMV 70
LP F+ N V LK + W + + +D I+ N W+ FVK + L +
Sbjct: 231 LPKTFSDNLKKKLPGN-VTLKGPSGVVWNVGLTARDDTIYFTNGWQRFVKDHSLKEN--- 398
Query: 71 WHLVLFEFKGPSKFEVCIF 89
++F++ G S FEV IF
Sbjct: 399 -DFLVFKYNGESLFEVLIF 452
>TC81461
Length = 745
Score = 45.8 bits (107), Expect = 3e-05
Identities = 24/75 (32%), Positives = 33/75 (44%)
Frame = +1
Query: 129 LPNGFMKKYSGGMSNPVFLKAPDNTSWEIHWSKHDSYFWFQKGWKEFVKYYSLDHGHLVL 188
LP F + V LK P W I + D +F GW++FV +SL +
Sbjct: 238 LPKTFSDNLKKKLPENVTLKGPSGVVWNIGLTTIDDTVYFTNGWQQFVNDHSLKESDFLF 417
Query: 189 FEFKDASHFGVHIFD 203
F++ S F V IFD
Sbjct: 418 FKYNGESLFEVLIFD 462
Score = 38.9 bits (89), Expect = 0.004
Identities = 24/80 (30%), Positives = 37/80 (46%)
Frame = +1
Query: 11 LPNAFTRKYSGDNMSNPVFLKPLDNISWEINWSKQDSDIWLQNVWEEFVKLYYLDQGHMV 70
LP F+ N V LK + W I + D ++ N W++FV + L + +
Sbjct: 238 LPKTFSDNLKKKLPEN-VTLKGPSGVVWNIGLTTIDDTVYFTNGWQQFVNDHSLKESDFL 414
Query: 71 WHLVLFEFKGPSKFEVCIFD 90
+ F++ G S FEV IFD
Sbjct: 415 F----FKYNGESLFEVLIFD 462
>TC80384
Length = 813
Score = 43.9 bits (102), Expect = 1e-04
Identities = 23/79 (29%), Positives = 42/79 (53%)
Frame = +2
Query: 318 PISFIREYLNEKKQTIKLKIEENLWPVEFIYYPEHGSGKLSKGWIQFAKESKLVGGDVCV 377
P +I +L Q + L++ +N W ++ Y+ +G L+ GW FA ++ L DVC+
Sbjct: 152 PNKWILNHLAPSSQDVILRMGKNEWLGKYCYHNIRNNGGLTGGWKYFALDNNLEEFDVCL 331
Query: 378 FELINKDEENPVLEVHIFK 396
F+ + +LE+ IF+
Sbjct: 332 FKPAGHLHDILILEMSIFR 388
>TC77100
Length = 926
Score = 42.4 bits (98), Expect = 4e-04
Identities = 29/84 (34%), Positives = 40/84 (47%), Gaps = 4/84 (4%)
Frame = +1
Query: 318 PISFIREYLNEKKQTIKLKIEENLWPVEFIYYPEHGSGKL----SKGWIQFAKESKLVGG 373
P S I E L K+ LK W + + +G K S W +FA+++ L G
Sbjct: 337 PSSSICEELPSKEVPTILKYRGKSWGMTY-----NGQNKTKQFDSVSWEKFAEDNYLKLG 501
Query: 374 DVCVFELINKDEENPVLEVHIFKG 397
D CVFEL+ EE V +V I +G
Sbjct: 502 DACVFELMKNSEEEIVFKVQILRG 573
>TC80302
Length = 886
Score = 41.6 bits (96), Expect = 6e-04
Identities = 45/163 (27%), Positives = 66/163 (39%), Gaps = 15/163 (9%)
Frame = +3
Query: 238 PKSPMS--LSRPCKKLRTGKSKDVGKSP----------ELRKRKKVQVEGGEPSSLGPHF 285
PK P +P ++ G++K G+ P +LR K E + S L
Sbjct: 84 PKQPTGQRTKKPANEVTPGQTKKRGRPPKEGNSREGALDLRASNKEHSEAAQ-SRLSSAK 260
Query: 286 TAQGEARKYTPKNPSFTVSLKPVIKLAYRP-PVPISFIREYL-NEKKQTIKLKIEENLWP 343
+ A+ +T P F +K R VP F +L N K + I ++ W
Sbjct: 261 DEKKLAQSFTSTFPYFVKIIKTFNVDGPRILNVPHQFSIAHLPNGKIKIILRNLKGEQWT 440
Query: 344 VEFIYYPE-HGSGKLSKGWIQFAKESKLVGGDVCVFELINKDE 385
+ H S L GW+ F + + + GDVCVFELIN E
Sbjct: 441 ANSVPRSRVHTSHTLCGGWMSFVRANNIKLGDVCVFELINDCE 569
>BF003537
Length = 397
Score = 40.4 bits (93), Expect = 0.001
Identities = 16/57 (28%), Positives = 28/57 (49%)
Frame = +3
Query: 120 PCLEAAELILPNGFMKKYSGGMSNPVFLKAPDNTSWEIHWSKHDSYFWFQKGWKEFV 176
P + L +P+GF+ + G V L P +W++ + D++ +F GW FV
Sbjct: 207 PSSTSQSLRVPDGFVHQMEGATCGLVSLTGPSGNTWQVRLVEQDNHLFFHHGWSTFV 377
>CA921493
Length = 656
Score = 39.7 bits (91), Expect = 0.002
Identities = 24/89 (26%), Positives = 43/89 (47%), Gaps = 2/89 (2%)
Frame = -1
Query: 317 VPISFIREYLNEKKQTIKLKIEEN-LWPVEFIYYPEHGS-GKLSKGWIQFAKESKLVGGD 374
+P F +L + K + L+ W V + Y + G GW+ F +++ + GD
Sbjct: 506 IPRRFSAAHLPDDKTEVTLRNSRGECWTVNSVPYAKRGMLHTFCGGWMSFVRDNGVNFGD 327
Query: 375 VCVFELINKDEENPVLEVHIFKGFKSRFN 403
C+FEL++ + V++VHI+ K N
Sbjct: 326 TCIFELVS----DYVMQVHIYGVGKESIN 252
>CA989827
Length = 345
Score = 36.6 bits (83), Expect = 0.020
Identities = 14/36 (38%), Positives = 25/36 (68%)
Frame = -2
Query: 360 GWIQFAKESKLVGGDVCVFELINKDEENPVLEVHIF 395
GW+ F +++ + GD C+FEL++ + V++VHIF
Sbjct: 263 GWVAFVRDNNIKFGDTCIFELVS----DYVMQVHIF 168
>TC87578 similar to PIR|T12113|T12113 transcription factor - fava bean,
partial (49%)
Length = 1021
Score = 36.2 bits (82), Expect = 0.026
Identities = 29/105 (27%), Positives = 48/105 (45%), Gaps = 5/105 (4%)
Frame = +2
Query: 212 PSNENQGEEHNPDQIGDEILLPKKTKPKSPMSLSRPCKKLRTGKSKDVGKSPELRKRKKV 271
P++++ ++ + Q GDE +P K +PK +S S+ T SK K + +KK
Sbjct: 584 PTDDSGADDSDASQSGDEKEIPAKKEPKKDLS-SKASASTSTSTSKKKSKDADEDGKKKK 760
Query: 272 QVEGGEPSS-----LGPHFTAQGEARKYTPKNPSFTVSLKPVIKL 311
Q + +P++ G F +Q E NP +S V KL
Sbjct: 761 QKKKKDPNAPKRGMSGFMFFSQMERENIKKANPG--ISFTDVAKL 889
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.138 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,647,215
Number of Sequences: 36976
Number of extensions: 282003
Number of successful extensions: 1453
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 1397
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1442
length of query: 441
length of database: 9,014,727
effective HSP length: 99
effective length of query: 342
effective length of database: 5,354,103
effective search space: 1831103226
effective search space used: 1831103226
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0040.3


