

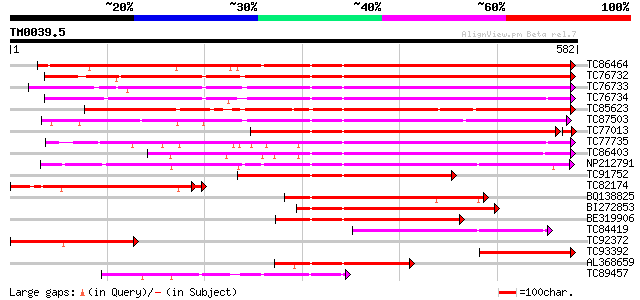
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0039.5
(582 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86464 similar to GP|2098711|gb|AAB57670.1| pectinesterase {Cit... 452 e-127
TC76732 homologue to PIR|T06468|T06468 pectinesterase (EC 3.1.1.... 445 e-125
TC76733 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11... 433 e-122
TC76734 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11... 417 e-117
TC85623 similar to GP|13605696|gb|AAK32841.1 At2g45220/F4L23.27 ... 414 e-116
TC87503 similar to SP|Q43111|PME3_PHAVU Pectinesterase 3 precurs... 407 e-114
TC77013 similar to GP|7025485|gb|AAF35897.1| pectin methylestera... 395 e-109
TC77735 similar to GP|14334646|gb|AAK59501.1 putative pectineste... 381 e-106
TC86403 similar to PIR|B86158|B86158 hypothetical protein AAF028... 366 e-101
NP212791 NP212791|AJ249611.1|CAB65291.1 pectin methyl-esterase PEF1 302 2e-82
TC91752 similar to GP|6630559|gb|AAF19578.1| putative pectineste... 247 1e-65
TC82174 similar to GP|4097684|gb|AAD00154.1| ubiquitin conjugati... 204 3e-57
BQ138825 similar to PIR|T49922|T49 pectin methylesterase-like pr... 196 3e-50
BI272853 weakly similar to PIR|T04359|T04 pectin methylesterase-... 191 5e-49
BE319906 similar to SP|Q43062|PME_ Pectinesterase PPE8B precurso... 187 7e-48
TC84419 similar to SP|Q42920|PME_MEDSA Pectinesterase precursor ... 168 6e-42
TC92372 weakly similar to GP|9759184|dbj|BAB09799.1 pectinestera... 152 3e-37
TC93392 similar to PIR|T49241|T49241 pectinesterase-like protein... 138 5e-33
AL368659 similar to PIR|B86158|B86 hypothetical protein AAF02886... 138 7e-33
TC89457 weakly similar to SP|Q43062|PME_PRUPE Pectinesterase PPE... 132 5e-31
>TC86464 similar to GP|2098711|gb|AAB57670.1| pectinesterase {Citrus
sinensis}, partial (85%)
Length = 1993
Score = 452 bits (1164), Expect = e-127
Identities = 252/583 (43%), Positives = 358/583 (61%), Gaps = 31/583 (5%)
Frame = +1
Query: 29 TTSGRKKKLIILVLLAVVLIVASAVVLTAVRSRAGGSGASTLRAKPTQAISN-------- 80
+ SG KKKL+I L +LIVAS V + A ++ + A + ++S+
Sbjct: 82 SNSGNKKKLLIS-LFTTLLIVASIVAIVATTTKNSNKSKNNSIASSSLSLSHHSHAILKS 258
Query: 81 TCSKTRFPSLCVKSLLDFPGST--AASERDLVHISLNMTFQHLAKALYS--SAAISASMD 136
C+ T +P LC ++ P T + +D++ +SLN+T + + ++ + S+
Sbjct: 259 ACTTTLYPELCFSAISSEPNITHKITNHKDVISLSLNITTRAVEHNYFTVEKLLLRKSLT 438
Query: 137 PRVRAAYDDCLELLDDSVDALGRSLSSFAAGSS-------SSDVMTWLSAALTNQDTCAD 189
R + A DCLE +D+++D L + + S + D+ T +S+A+TNQ TC D
Sbjct: 439 KREKIALHDCLETIDETLDELKEAQNDLVLYPSKKTLYQHADDLKTLISSAITNQVTCLD 618
Query: 190 GFA--DTSGDVKDQMAGNLKDLSELVSNCLAIFAGAGD----DFSGVPI-----QNRRLL 238
GF+ D +V+ + + + SN LA+ D +F + +NR+LL
Sbjct: 619 GFSHDDADKEVRKVLQEGQIHVEHMCSNALAMTKNMTDKDIAEFEQTNMVLGSNKNRKLL 798
Query: 239 AMRDD-NFPRWLNRRDRRLLSLPVSAMQADVVVSKDGNGTAKTIAEAIKKVPEYGSRRFI 297
+ +P W++ DRRLL S ++ADVVV+ DG+G KT++EA+ P S+R++
Sbjct: 799 EEENGVGWPEWISAGDRRLLQ--GSTVKADVVVAADGSGNFKTVSEAVAAAPLKSSKRYV 972
Query: 298 IYVRAGRYEENNLKIGRKRTNVMIIGDGKGKTVITGGKNVMDGLLTTFHTASFAASGAGF 357
I ++AG Y+EN +++ +K+TN+M +GDG+ T+ITG +NV+DG TTFH+A+ A G F
Sbjct: 973 IKIKAGVYKEN-VEVPKKKTNIMFLGDGRTNTIITGSRNVVDGS-TTFHSATVAIVGGNF 1146
Query: 358 IARDMTFENYAGPEKHQAVALRVGADHAVVYRCNVIGYQDTMYVHSNRQFYRECDIYGTV 417
+ARD+TF+N AGP KHQAVALRVGAD + Y C++I YQDT+YVH+NRQF+ C I GTV
Sbjct: 1147LARDITFQNTAGPAKHQAVALRVGADLSAFYNCDIIAYQDTLYVHNNRQFFVNCFISGTV 1326
Query: 418 DFIFGNAAVVFQNCSLYARKPMAQQKNTITAQNRKDPNQNTGISIHACRVLAAADLKASN 477
DFIFGN+AVVFQNC ++AR+P + QKN +TAQ R DPNQNTGI I CR+ A DL+
Sbjct: 1327DFIFGNSAVVFQNCDIHARRPNSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATKDLEGVK 1506
Query: 478 ESFETYLGRPWKLYSRTVYMMSYIGDHVHQRGWLEWNTTFALDTLYYGEYMNYGPGAAVG 537
+F TYLGRPWK YSRTV+M S I D + GW EWN FAL+TL Y EY N GPGA
Sbjct: 1507GNFPTYLGRPWKEYSRTVFMQSSISDVIDPVGWHEWNGNFALNTLVYREYQNTGPGAGTS 1686
Query: 538 QRVNWPGYRVITSALEASRFTVAQFILGATWLPSTGVAFLSGL 580
+RV W G++VITSA EA T FI G++WL STG F GL
Sbjct: 1687KRVTWKGFKVITSAAEAQSSTPGNFIGGSSWLGSTGFPFSLGL 1815
>TC76732 homologue to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11)
precursor - garden pea, complete
Length = 1860
Score = 445 bits (1144), Expect = e-125
Identities = 250/556 (44%), Positives = 341/556 (60%), Gaps = 11/556 (1%)
Frame = +2
Query: 36 KLIILVLLAVVLIVASAVVLTAVRSRAGGSGASTLRAKPTQAISNTCSKTRFPSLCVKSL 95
K LVL V +I +SA++++ + S+ N C C+ +
Sbjct: 83 KTFWLVLSLVAIISSSALIISHLNKPISFFNLSSA--------PNLCEHALDTKSCLTHV 238
Query: 96 LDF-PGSTAASERD-----LVHISLNMTFQHLAKALYSSAAISASMD-PRVRAAYDDCLE 148
+ GST ++ +D LV + L + H+ KA+ ++ I ++ PR A +DC E
Sbjct: 239 SEVVQGSTLSNTKDHKLSTLVSL-LTKSTAHIRKAMDTANVIKRRVNSPREEIALNDCEE 415
Query: 149 LLDDSVDALGRSLSSFAAGS--SSSDVMTWLSAALTNQDTCADGFADTSGDVKDQMAGNL 206
L+D S+D + S+ + + S D TWLS+ LTN TC DG +S V M +L
Sbjct: 416 LMDLSMDRVWDSVLTLTKNNIDSQHDAHTWLSSVLTNHATCLDGLEGSSRVV---MESDL 586
Query: 207 KDLSELVSNCLAIFAGAGDDFSGVPIQNRRLLAMRDDNFPRWLNRRDRRLLSLPVSAMQA 266
DL + LA+ + + +L + +FP W+ +DRRLL V ++A
Sbjct: 587 HDLISRARSSLAVLVSVLPPKANDGFIDEKL----NGDFPSWVTSKDRRLLESSVGDIKA 754
Query: 267 DVVVSKDGNGTAKTIAEAIKKVPEYGSRRFIIYVRAGRYEENNLKIGRKRTNVMIIGDGK 326
+VVV++DG+G KT+A+A+ P+ G R++IYV+ G Y+EN ++IG+K+TNVM++GDG
Sbjct: 755 NVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYVKKGTYKEN-IEIGKKKTNVMLVGDGM 931
Query: 327 GKTVITGGKNVMDGLLTTFHTASFAASGAGFIARDMTFENYAGPEKHQAVALRVGADHAV 386
T+ITG N +DG TTF +A+ AA G GFIA+D+ F+N AGP+KHQAVALRVGAD +V
Sbjct: 932 DATIITGSLNFIDGT-TTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAVALRVGADQSV 1108
Query: 387 VYRCNVIGYQDTMYVHSNRQFYRECDIYGTVDFIFGNAAVVFQNCSLYARKPMAQQKNTI 446
+ RC + +QDT+Y HSNRQFYR+ I GTVDFIFGNAAVVFQ L ARKPMA QKN +
Sbjct: 1109INRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAARKPMANQKNMV 1288
Query: 447 TAQNRKDPNQNTGISIHACRVLAAADLKASNESFETYLGRPWKLYSRTVYMMSYIGDHVH 506
TAQ R+DPNQNT SI C V+ ++DLK S +TYLGRPWK YSRTV + S + H+
Sbjct: 1289TAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKKYSRTVVLQSVVDGHID 1468
Query: 507 QRGWLEWNTTFA--LDTLYYGEYMNYGPGAAVGQRVNWPGYRVITSALEASRFTVAQFIL 564
GW EW+ L TLYYGEYMN G GA G+RV WPGY +I +A EAS+FTV Q I
Sbjct: 1469PAGWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAAEASKFTVTQLIQ 1648
Query: 565 GATWLPSTGVAFLSGL 580
G WL +TGVAF+ GL
Sbjct: 1649GNVWLKNTGVAFIEGL 1696
>TC76733 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11) precursor
- garden pea, partial (98%)
Length = 2029
Score = 433 bits (1114), Expect = e-122
Identities = 243/571 (42%), Positives = 343/571 (59%), Gaps = 10/571 (1%)
Frame = +2
Query: 20 VALVNSRTKTTSGRKK--KLIILVLLAVVLIVASAVVLTAVRSRAGGSGASTLRAKPTQA 77
+ + N ++ RK K L+L V +I++SA++ T ++ +T++ +
Sbjct: 41 ITMANQQSLLDKPRKSLPKTFWLILSLVAIIISSALISTHLKKPISFFHLTTVQNVYCEH 220
Query: 78 ISNTCSKTRFPSLCVKSLLDFPGSTAASERDLVHISLNMTFQ---HLAKALYSSAAISAS 134
+T S V + P +++L H+ L++ + H+ A+ +++ I
Sbjct: 221 AVDTKSCLAH----VSEVSHVPTLVTTKDQNL-HVLLSLLTKSTTHIQNAMDTASVIKRR 385
Query: 135 MD-PRVRAAYDDCLELLDDSVDALGRSLSSFAAGS--SSSDVMTWLSAALTNQDTCADGF 191
++ PR A DC +L+D S++ + ++ + S D TWLS+ LTN TC DG
Sbjct: 386 INSPREEIALSDCEQLMDLSMNRIWDTMLKLTKNNIDSQQDAHTWLSSVLTNHATCLDGL 565
Query: 192 ADTSGDVKDQMAGNLKDLSELVSNCLAIFAGAGDDFSGVPIQNRRLLAMRDDNFPRWLNR 251
+S V M +L+DL + LA+F + L+ FP W+
Sbjct: 566 EGSSRVV---MENDLQDLISRARSSLAVFLVVFPQKDRDQFIDETLIG----EFPSWVTS 724
Query: 252 RDRRLLSLPVSAMQADVVVSKDGNGTAKTIAEAIKKVPEYGSRRFIIYVRAGRYEENNLK 311
+DRRLL V ++A+VVV++DG+G KT+AEA+ P+ G +++IYV+ G Y+EN ++
Sbjct: 725 KDRRLLETAVGDIKANVVVAQDGSGKFKTVAEAVASAPDNGKTKYVIYVKKGTYKEN-VE 901
Query: 312 IGRKRTNVMIIGDGKGKTVITGGKNVMDGLLTTFHTASFAASGAGFIARDMTFENYAGPE 371
IG K+TNVM++GDG T+ITG N +DG TTF +++ AA G GFIA+D+ F+N AG
Sbjct: 902 IGSKKTNVMLVGDGMDATIITGNLNFIDGT-TTFKSSTVAAVGDGFIAQDIWFQNMAGAA 1078
Query: 372 KHQAVALRVGADHAVVYRCNVIGYQDTMYVHSNRQFYRECDIYGTVDFIFGNAAVVFQNC 431
KHQAVALRVG+D +V+ RC + +QDT+Y HSNRQFYR+ I GT+DFIFGNAAVVFQ C
Sbjct: 1079KHQAVALRVGSDQSVINRCRIDAFQDTLYAHSNRQFYRDSVITGTIDFIFGNAAVVFQKC 1258
Query: 432 SLYARKPMAQQKNTITAQNRKDPNQNTGISIHACRVLAAADLKASNESFETYLGRPWKLY 491
L ARKPMA Q N TAQ R+DP QNTG SI C + ++DLK S +T+LGRPWK Y
Sbjct: 1259KLVARKPMANQNNMFTAQGREDPGQNTGTSIQQCDLTPSSDLKPVVGSIKTFLGRPWKKY 1438
Query: 492 SRTVYMMSYIGDHVHQRGWLEWNTTFA--LDTLYYGEYMNYGPGAAVGQRVNWPGYRVIT 549
SRTV M S++ H+ GW EW+ L TLYYGEY+N GPGA +RV WPGY VI
Sbjct: 1439SRTVVMQSFLDSHIDPTGWAEWDAASKDFLQTLYYGEYLNNGPGAGTAKRVTWPGYHVIN 1618
Query: 550 SALEASRFTVAQFILGATWLPSTGVAFLSGL 580
+A EAS+FTVAQ I G WL +TGVAF GL
Sbjct: 1619TAAEASKFTVAQLIQGNVWLKNTGVAFTEGL 1711
>TC76734 similar to PIR|T06468|T06468 pectinesterase (EC 3.1.1.11) precursor
- garden pea, complete
Length = 1981
Score = 417 bits (1072), Expect = e-117
Identities = 239/555 (43%), Positives = 329/555 (59%), Gaps = 10/555 (1%)
Frame = +3
Query: 36 KLIILVLLAVVLIVASAVVLTAVRSRAGGSGASTLRAKPTQAISNTCSKTRFPSLCVKSL 95
K LVL +I +SA++++ + S+ A+ T + S
Sbjct: 90 KTFWLVLSLAAIISSSALIISHLNKPISIFHFSSAPNVCEHAVDTNSCLTHVAEVVQGST 269
Query: 96 LDFPGSTAASERDLVHISLNMTFQHLAKALYSSAAISASMD-PRVRAAYDDCLELLDDSV 154
LD +T + + L + H+ KA+ ++ I ++ PR A + C +L++ S+
Sbjct: 270 LD---NTKDHKLSTLISLLTKSTTHIRKAMDTANVIKRRINSPREENALNVCEKLMNLSM 440
Query: 155 DALGRSLSSFAAGS--SSSDVMTWLSAALTNQDTCADGFADTSGDVKDQMAGNLKDLSEL 212
+ + S+ + + S D TWLS+ LTN TC DG TS V M +++DL
Sbjct: 441 ERVWDSVLTLTKDNMDSQQDAHTWLSSVLTNHATCLDGLEGTSRAV---MENDIQDLIAR 611
Query: 213 VSNCLAIFAGA-----GDDFSGVPIQNRRLLAMRDDNFPRWLNRRDRRLLSLPVSAMQAD 267
+ LA+ D+F + +FP W+ +DRRLL V ++A+
Sbjct: 612 ARSSLAVLVAVLPPKDHDEFIDESLNG---------DFPSWVTSKDRRLLESSVGDVKAN 764
Query: 268 VVVSKDGNGTAKTIAEAIKKVPEYGSRRFIIYVRAGRYEENNLKIGRKRTNVMIIGDGKG 327
VVV+KDG+G KT+AEA+ P G+ R++IYV+ G Y+EN ++I +TNVM++GDG
Sbjct: 765 VVVAKDGSGKFKTVAEAVASAPNKGTARYVIYVKKGIYKEN-VEIASSKTNVMLLGDGMD 941
Query: 328 KTVITGGKNVMDGLLTTFHTASFAASGAGFIARDMTFENYAGPEKHQAVALRVGADHAVV 387
T+ITG N +DG TF TA+ AA G FIA+D+ F+N AGP+KHQAVALRVG+D +V+
Sbjct: 942 ATIITGSLNYVDGT-GTFQTATVAAVGDWFIAQDIGFQNTAGPQKHQAVALRVGSDRSVI 1118
Query: 388 YRCNVIGYQDTMYVHSNRQFYRECDIYGTVDFIFGNAAVVFQNCSLYARKPMAQQKNTIT 447
RC + +QDT+Y H+NRQFYR+ I GT+DFIFG+AAVV Q C L ARKPMA Q N +T
Sbjct: 1119NRCKIDAFQDTLYAHTNRQFYRDSFITGTIDFIFGDAAVVLQKCKLVARKPMANQNNMVT 1298
Query: 448 AQNRKDPNQNTGISIHACRVLAAADLKASNESFETYLGRPWKLYSRTVYMMSYIGDHVHQ 507
AQ R DPNQNT SI C V+ + DLK S +TYLGRPWK YSRTV M S +G H+
Sbjct: 1299AQGRIDPNQNTATSIQQCDVIPSTDLKPVIGSVKTYLGRPWKKYSRTVVMQSLLGAHIDP 1478
Query: 508 RGWLEWNTTFA--LDTLYYGEYMNYGPGAAVGQRVNWPGYRVITSALEASRFTVAQFILG 565
GW EW+ L TLYYGEYMN GPGA +RV WPGY +I +A EA++FTVAQ I G
Sbjct: 1479TGWAEWDAASKDFLQTLYYGEYMNSGPGAGTSKRVKWPGYHIINTA-EANKFTVAQLIQG 1655
Query: 566 ATWLPSTGVAFLSGL 580
WL +TGVAF++GL
Sbjct: 1656NVWLKNTGVAFIAGL 1700
>TC85623 similar to GP|13605696|gb|AAK32841.1 At2g45220/F4L23.27 {Arabidopsis
thaliana}, partial (79%)
Length = 2442
Score = 414 bits (1063), Expect = e-116
Identities = 236/508 (46%), Positives = 317/508 (61%), Gaps = 4/508 (0%)
Frame = +3
Query: 77 AISNTCSKTRFPSLCVKSLLDFP-GSTAASERDLVHISLNMTFQHLAKALYSSAAISASM 135
+I + CS+T +P C L + T S+ D +SL + + K ++ A+
Sbjct: 648 SIKSWCSQTPYPQPCEYYLTNNAFNQTIKSKSDFFKVSLEIAMERAKKGEENTHAVGPKC 827
Query: 136 -DPRVRAAYDDCLELLDDSVDALGRSLSSFAAGSSSSDVMTWLSAALTNQDTCADGFAD- 193
P+ +AA+ DCLEL D +V L ++ + S TWLS ALTN +TC +GF D
Sbjct: 828 RSPQEKAAWADCLELYDFTVQKLSQTKYTKCTQYESQ---TWLSTALTNLETCKNGFYDL 998
Query: 194 -TSGDVKDQMAGNLKDLSELVSNCLAIFAGAGDDFSGVPIQNRRLLAMRDDNFPRWLNRR 252
+ V ++ N+ ++L+SN L++ + VP Q D FP W+
Sbjct: 999 GVTNYVLPLLSNNV---TKLLSNTLSL--------NKVPYQQPSY----KDGFPTWVKPG 1133
Query: 253 DRRLLSLPVSAMQADVVVSKDGNGTAKTIAEAIKKVPEYGSRRFIIYVRAGRYEENNLKI 312
DR+LL +A +A+VVV+KDG+G T+ A P GS R++IYV+AG Y E ++
Sbjct: 1134 DRKLLQTSSAASKANVVVAKDGSGKYTTVKAATDAAPS-GSGRYVIYVKAGVYNE---QV 1301
Query: 313 GRKRTNVMIIGDGKGKTVITGGKNVMDGLLTTFHTASFAASGAGFIARDMTFENYAGPEK 372
K NVM++GDG GKT+ITG K+V G TTF +A+ AA+G GFIA+D+TF N AG
Sbjct: 1302 EIKAKNVMLVGDGIGKTIITGSKSVGGGT-TTFRSATVAATGDGFIAQDITFRNTAGAAN 1478
Query: 373 HQAVALRVGADHAVVYRCNVIGYQDTMYVHSNRQFYRECDIYGTVDFIFGNAAVVFQNCS 432
HQAVA R G+D +V Y+C+ GYQDT+YVHS RQFYREC+IYGTVDFIFGNAAVV QNC+
Sbjct: 1479 HQAVAFRSGSDLSVFYKCSFEGYQDTLYVHSERQFYRECNIYGTVDFIFGNAAVVLQNCN 1658
Query: 433 LYARKPMAQQKNTITAQNRKDPNQNTGISIHACRVLAAADLKASNESFETYLGRPWKLYS 492
++AR P A + T+TAQ R DPNQNTGI IH RV A +DL S S ++YLGRPW+ YS
Sbjct: 1659 IFARNPPA-KTITVTAQGRTDPNQNTGIIIHNSRVSAQSDLNPS--SVKSYLGRPWQKYS 1829
Query: 493 RTVYMMSYIGDHVHQRGWLEWNTTFALDTLYYGEYMNYGPGAAVGQRVNWPGYRVITSAL 552
RTV+M + + ++ GWL W+ FALDTLYY EY N G G++ RV W GY V+TSA
Sbjct: 1830 RTVFMKTVLDGFINPAGWLPWDGNFALDTLYYAEYANTGSGSSTSNRVTWKGYHVLTSAS 2009
Query: 553 EASRFTVAQFILGATWLPSTGVAFLSGL 580
+AS FTV FI G +W+ +TGV F SGL
Sbjct: 2010 QASPFTVGNFIAGNSWIGNTGVPFTSGL 2093
>TC87503 similar to SP|Q43111|PME3_PHAVU Pectinesterase 3 precursor (EC
3.1.1.11) (Pectin methylesterase 3) (PE 3)., partial
(93%)
Length = 2088
Score = 407 bits (1046), Expect = e-114
Identities = 236/563 (41%), Positives = 318/563 (55%), Gaps = 19/563 (3%)
Frame = +2
Query: 33 RKKKLIILV---LLAVVLIVASAVVLTAVRSRAGGSGASTL---RAKPTQAISNTCSKTR 86
RK+ II++ +L V+I A A +L + S ++L P ++ C T+
Sbjct: 134 RKRITIIIISSIILVAVIIAAVAGILIHKHNTESSSSPNSLPNTELTPATSLKAVCESTQ 313
Query: 87 FPSLCVKSLLDFPGSTAASERDLVHISLNMTFQHLAKALYSSAAISASMDPRVRAAYDDC 146
+P+ C S+ P S L +SL + L+K + + A+ +PRV+ A C
Sbjct: 314 YPNSCFSSISSLPDSNTTDPEQLFKLSLKVAIDELSKLSLTRFSEKAT-EPRVKKAIGVC 490
Query: 147 LELLDDSVDALGRSLSSFAAGSSS------SDVMTWLSAALTNQDTCADGFADTSGD--- 197
+L DS+D L S+S+ G DV TWLSAALT+ DTC D + +
Sbjct: 491 DNVLADSLDRLNDSMSTIVDGGKMLSPAKIRDVETWLSAALTDHDTCLDAVGEVNSTAAR 670
Query: 198 -VKDQMAGNLKDLSELVSNCLAIFAGAGDDFSGVPIQN--RRLLAMRDDNFPRWLNRRDR 254
V ++ +++ +E SN LAI + S + N RRLL FP WL +R
Sbjct: 671 GVIPEIERIMRNSTEFASNSLAIVSKVIRLLSNFEVSNHHRRLLG----EFPEWLGTAER 838
Query: 255 RLLSLPVSAMQADVVVSKDGNGTAKTIAEAIKKVPEYGSRRFIIYVRAGRYEENNLKIGR 314
RLL+ V+ D VV+KDG+G KTI EA+K V + +RF++YV+ G Y EN + + +
Sbjct: 839 RLLATVVNETVPDAVVAKDGSGQYKTIGEALKLVKKKSLQRFVVYVKKGVYVEN-IDLDK 1015
Query: 315 KRTNVMIIGDGKGKTVITGGKNVMDGLLTTFHTASFAASGAGFIARDMTFENYAGPEKHQ 374
NVMI GDG +TV++G +N +DG TF TA+FA G GFIA+D+ F N AG KHQ
Sbjct: 1016NTWNVMIYGDGMTETVVSGSRNYIDGT-PTFETATFAVKGKGFIAKDIQFLNTAGASKHQ 1192
Query: 375 AVALRVGADHAVVYRCNVIGYQDTMYVHSNRQFYRECDIYGTVDFIFGNAAVVFQNCSLY 434
AVA+R G+D +V YRC+ +GYQDT+Y HSNRQFYR+CDI GT+DFIFGNAA VFQNC +
Sbjct: 1193AVAMRSGSDQSVFYRCSFVGYQDTLYAHSNRQFYRDCDITGTIDFIFGNAAAVFQNCKIM 1372
Query: 435 ARKPMAQQKNTITAQNRKDPNQNTGISIHACRVLAAADLKASNESFETYLGRPWKLYSRT 494
R+PM+ Q NTITAQ +KDPNQN+GI I L N TYLGRPWK +S T
Sbjct: 1373PRQPMSNQFNTITAQGKKDPNQNSGIVIQKS---TFTTLPGDNLIAPTYLGRPWKDFSTT 1543
Query: 495 VYMMSYIGDHVHQRGWLEWNTTF-ALDTLYYGEYMNYGPGAAVGQRVNWPGYRVITSALE 553
+ M S IG + GW+ W ++ Y EY N GPGA V RV W GY+ +
Sbjct: 1544IIMKSEIGSFLKPVGWISWVANVEPPSSILYAEYQNTGPGADVPGRVKWAGYKPALGDED 1723
Query: 554 ASRFTVAQFILGATWLPSTGVAF 576
A +FTV FI G WLPS V F
Sbjct: 1724AIKFTVDSFIQGPEWLPSASVQF 1792
>TC77013 similar to GP|7025485|gb|AAF35897.1| pectin methylesterase isoform
alpha {Vigna radiata}, partial (94%)
Length = 1655
Score = 395 bits (1014), Expect(2) = e-109
Identities = 189/318 (59%), Positives = 243/318 (75%)
Frame = +2
Query: 248 WLNRRDRRLLSLPVSAMQADVVVSKDGNGTAKTIAEAIKKVPEYGSRRFIIYVRAGRYEE 307
W++ +DR+LL V+ + D+VV+KDG G TI EAI P + RF+I+++AG Y E
Sbjct: 41 WVSPKDRKLLQAAVNQTKFDLVVAKDGTGNFATIGEAIAAAPNSSATRFVIHIKAGAYFE 220
Query: 308 NNLKIGRKRTNVMIIGDGKGKTVITGGKNVMDGLLTTFHTASFAASGAGFIARDMTFENY 367
N +++ +K+TN+M++GDG G+TV+ +NV+DG TTF +++FA G FIA+ +TFEN
Sbjct: 221 N-VEVIKKKTNLMLVGDGIGQTVVKASRNVVDGW-TTFQSSTFAVVGDKFIAKGITFENS 394
Query: 368 AGPEKHQAVALRVGADHAVVYRCNVIGYQDTMYVHSNRQFYRECDIYGTVDFIFGNAAVV 427
AGP KHQAVA+R GAD + Y+C+ + YQDT+YVHS RQFYRECD+YGTVDFIFGNAAVV
Sbjct: 395 AGPSKHQAVAVRNGADFSAFYQCSFVAYQDTLYVHSLRQFYRECDVYGTVDFIFGNAAVV 574
Query: 428 FQNCSLYARKPMAQQKNTITAQNRKDPNQNTGISIHACRVLAAADLKASNESFETYLGRP 487
FQNC+LYARKP +QKN TAQ R+DPNQNTGISI C++ AAADL +F++YLGRP
Sbjct: 575 FQNCNLYARKPDPKQKNLFTAQGREDPNQNTGISILNCKIAAAADLIPVQSTFKSYLGRP 754
Query: 488 WKLYSRTVYMMSYIGDHVHQRGWLEWNTTFALDTLYYGEYMNYGPGAAVGQRVNWPGYRV 547
WK YSRTVY+ S+I + + GWLEWN TFALDTL+YGEY N GPG+ RV WPGY+V
Sbjct: 755 WKKYSRTVYLNSFIDNLIDPPGWLEWNGTFALDTLFYGEYKNRGPGSNTSARVTWPGYKV 934
Query: 548 ITSALEASRFTVAQFILG 565
IT+A EAS+FTV QFI G
Sbjct: 935 ITNATEASQFTVRQFIQG 988
Score = 20.8 bits (42), Expect(2) = e-109
Identities = 9/14 (64%), Positives = 9/14 (64%)
Frame = +1
Query: 568 WLPSTGVAFLSGLS 581
WL STGV F LS
Sbjct: 997 WLNSTGVPFFLDLS 1038
>TC77735 similar to GP|14334646|gb|AAK59501.1 putative pectinesterase
{Arabidopsis thaliana}, partial (59%)
Length = 1963
Score = 381 bits (978), Expect = e-106
Identities = 234/586 (39%), Positives = 323/586 (54%), Gaps = 42/586 (7%)
Frame = +2
Query: 37 LIILVLLAVVLIVASAVVLTAVRSRAGGSGASTLRAKPTQAISNTCSKTRFPSLCVKSLL 96
L++ +LLA +L++A S + P + S C T +P LC +S+L
Sbjct: 59 LLLFILLAPLLLLAQ-------------SPPPSPSPPPPSSSSAACKTTLYPKLC-RSML 196
Query: 97 DFPGSTAASERDLVHISLNMTFQHLAKA------LYSSAAISASMDPRVRAAYDDCLELL 150
S+ + + S+ + K + S+S++ A DC +L
Sbjct: 197 SAIRSSPSDPYNYGKFSIKQNLKVARKLEKVFIDFLNRHQSSSSLNHEEVGALVDCKDLN 376
Query: 151 DDSVD---ALGRSLSSFAAGSSSSD------VMTWLSAALTNQDTCADGFADTSGDVKDQ 201
+VD ++ L S ++ SSSSD + ++LSA TN TC DG T ++ +
Sbjct: 377 SLNVDYLESISDELKSASSSSSSSDTELVDKIESYLSAVATNHYTCYDGLVVTKSNIANA 556
Query: 202 MAGNLKDLSELVSNCLAIFAGAGDDFS--------GVPIQN-------RRLLAMRDDNFP 246
+A LKD ++ S L + A G+P ++ +L+ + +
Sbjct: 557 LAVPLKDATQFYSVSLGLVTEALSKNMKRNKTRKHGLPNKSFKVRQPLEKLIKLLRTKYS 736
Query: 247 ------RWLNRRDRRLLSLPVS---AMQADVVVSKDGNGTAKTIAEAIKKVPEYGSRR-- 295
+ R R+L S + V+VS G +I +AI P
Sbjct: 737 CQKTSSNCTSTRTERILKESESHGILLNDFVLVSPYGIANHTSIGDAIAAAPNNTKPEDG 916
Query: 296 -FIIYVRAGRYEENNLKIGRKRTNVMIIGDGKGKTVITGGKNVMDGLLTTFHTASFAASG 354
++IYVR G YEE + + + + N++++GDG T+ITG +V+DG TTF++++FA SG
Sbjct: 917 YYLIYVREGYYEEYVI-VPKHKNNILLVGDGINNTIITGNHSVIDGW-TTFNSSTFAVSG 1090
Query: 355 AGFIARDMTFENYAGPEKHQAVALRVGADHAVVYRCNVIGYQDTMYVHSNRQFYRECDIY 414
FIA D+TF N AGPEKHQAVA+R AD + YRC+ GYQDT+YVHS RQFYR+C IY
Sbjct: 1091ERFIAVDITFRNTAGPEKHQAVAVRNNADLSTFYRCSFEGYQDTLYVHSVRQFYRDCKIY 1270
Query: 415 GTVDFIFGNAAVVFQNCSLYARKPMAQQKNTITAQNRKDPNQNTGISIHACRVLAAADLK 474
GTVDFIFGNAAVVFQNC++YARKP+ QKN +TAQ R DPNQNTGISI C + AA DL
Sbjct: 1271GTVDFIFGNAAVVFQNCNIYARKPLPNQKNAVTAQGRTDPNQNTGISIQNCTIDAAQDLA 1450
Query: 475 ASNESFETYLGRPWKLYSRTVYMMSYIGDHVHQRGWLEWNTTFALDTLYYGEYMNYGPGA 534
S +YLGRPWK+YSRTVYM SYIGD V GWLEWN T LDT++YGE+ NYGPG+
Sbjct: 1451NDLNSTMSYLGRPWKIYSRTVYMQSYIGDFVQPSGWLEWNGTVGLDTIFYGEFNNYGPGS 1630
Query: 535 AVGQRVNWPGYRVITSALEASRFTVAQFILGATWLPSTGVAFLSGL 580
RV WPG+ ++ +A FTV F LG TWLP T + + GL
Sbjct: 1631VTNNRVQWPGHFLLNDT-QAWNFTVLNFTLGNTWLPDTDIPYTEGL 1765
>TC86403 similar to PIR|B86158|B86158 hypothetical protein AAF02886.1
[imported] - Arabidopsis thaliana, partial (63%)
Length = 1929
Score = 366 bits (939), Expect = e-101
Identities = 213/467 (45%), Positives = 273/467 (57%), Gaps = 28/467 (5%)
Frame = +3
Query: 142 AYDDCLELLDDSVDALGRSLSS------FAAGSSSSDVMTWLSAALTNQDTCADGFADTS 195
A DC L D + D L S + F ++ T LSA LTNQ TC DG DTS
Sbjct: 315 ALQDCQSLSDLNFDFLSSSFQTVNKTTKFLPSLQGENIQTLLSAILTNQQTCLDGLKDTS 494
Query: 196 G--DVKDQMAGNLKDLSELVSNCLAIFA----GAGDDFSGVPIQNRRLLAMRDDNFPRWL 249
++ + L + ++L S LA F + + P ++ P +
Sbjct: 495 SAWSFRNGLTIPLSNDTKLYSVSLAFFTKGWVNPKTNKTSFPNSKHSNKGFKNGRLPLKM 674
Query: 250 NRRDRRLLS-------LPVSAMQADVVV------SKDGNGTAKTIAEAIKKVPEYGSRR- 295
+ R + L + + DVVV S+DG+G TI +AI P
Sbjct: 675 TSKTRAIYESVSRRKLLQSNQVGEDVVVRDIVTVSQDGSGNFTTINDAIAAAPNKSVSSD 854
Query: 296 --FIIYVRAGRYEENNLKIGRKRTNVMIIGDGKGKTVITGGKNVMDGLLTTFHTASFAAS 353
F+IYV AG YEE + I +K+T +M+IGDG KT+ITG +V+DG TTF + +FA
Sbjct: 855 GYFLIYVTAGVYEEY-ITIDKKKTYLMMIGDGINKTIITGNHSVVDGW-TTFGSPTFAVV 1028
Query: 354 GAGFIARDMTFENYAGPEKHQAVALRVGADHAVVYRCNVIGYQDTMYVHSNRQFYRECDI 413
G GF+ +MT N AG KHQAVALR GAD + Y C+ GYQDT+Y HS RQFYRECDI
Sbjct: 1029GQGFVGVNMTIRNTAGAVKHQAVALRNGADLSTFYSCSFEGYQDTLYTHSLRQFYRECDI 1208
Query: 414 YGTVDFIFGNAAVVFQNCSLYARKPMAQQKNTITAQNRKDPNQNTGISIHACRVLAAADL 473
YGTVDFIFGNA VVFQNC+LY R PM+ Q N ITAQ R DPNQ+TG SIH C + A DL
Sbjct: 1209YGTVDFIFGNAKVVFQNCNLYPRLPMSGQFNAITAQGRTDPNQDTGTSIHNCTIKATDDL 1388
Query: 474 KASNESFETYLGRPWKLYSRTVYMMSYIGDHVHQRGWLEWNTTFALDTLYYGEYMNYGPG 533
ASN + TYLGRPWK YSRTVYM +++ + ++ GW W+ FAL TLYY E+ N GPG
Sbjct: 1389AASNGAVSTYLGRPWKEYSRTVYMQTFMDNVINVAGWRAWDGEFALSTLYYAEFNNSGPG 1568
Query: 534 AAVGQRVNWPGYRVITSALEASRFTVAQFILGATWLPSTGVAFLSGL 580
++ RV W GY VI +A +A+ FTVA F+LG WLP TGV++ + L
Sbjct: 1569SSTDGRVTWQGYHVI-NATDAANFTVANFLLGDDWLPQTGVSYTNSL 1706
>NP212791 NP212791|AJ249611.1|CAB65291.1 pectin methyl-esterase PEF1
Length = 1698
Score = 302 bits (774), Expect = 2e-82
Identities = 198/565 (35%), Positives = 294/565 (51%), Gaps = 17/565 (3%)
Frame = +1
Query: 32 GRKKKLIILVLLAVVLIVASAVVLTAVRSRAGGSGASTLRAKPTQA-ISNTCSKTRFPSL 90
G+ KK +L + ++L+ VV AV GG G +Q + C T+F
Sbjct: 31 GQGKKHALLGVSCILLVAMVGVV--AVSLTKGGDGEQKAHISNSQKNVDMLCQSTKFKET 204
Query: 91 CVKSLLDFPGSTAASERDLVHISLNMTFQHLAKALYSSAAISA-SMDPRVRAAYDDCLEL 149
C K+L ++ ++ ++ + +L T + L K + +SA + D + A + C E+
Sbjct: 205 CHKTL---EKASFSNMKNRIKGALGATEEELRKHINNSALYQELATDSMTKQAMEICNEV 375
Query: 150 LDDSVDALGRSLSSF------AAGSSSSDVMTWLSAALTNQDTCADGFADTSGDVKDQMA 203
LD +VD + +S+ + + D+ WL+ L++Q TC DGF +T + MA
Sbjct: 376 LDYAVDGIHKSVGTLDQFDFHKLSEYAFDIKVWLTGTLSHQQTCLDGFVNTKTHAGETMA 555
Query: 204 GNLKDLSELVSNCLAIFAGAGDDFSGV-PIQ---NRRLLAMRDDNFPRWLNRRDRRLLSL 259
LK EL SN + + G P Q +RRLL+ DD P W++ R LL+
Sbjct: 556 KVLKTSMELSSNAIDMMDVVSRILKGFHPSQYGVSRRLLS--DDGIPSWVSDGHRHLLA- 726
Query: 260 PVSAMQADVVVSKDGNGTAKTIAEAIKKVPEYGSRRFIIYVRAGRYEENNLKIGRKRTNV 319
++A+ VV++DG+G KT+ +A+K VP + F+IYV+AG Y+E + + ++ V
Sbjct: 727 -GGNVKANAVVAQDGSGQFKTLTDALKTVPPTNAAPFVIYVKAGVYKET-VNVAKEMNYV 900
Query: 320 MIIGDGKGKTVITGGKNVMDGLLTTFHTASFAASGAGFIARDMTFENYAGPEKHQAVALR 379
+IGDG KT TG N DG+ T+ TA+F +GA F+A+D+ FEN AG K QAVALR
Sbjct: 901 TVIGDGPTKTKFTGSLNYADGI-NTYKTATFGVNGANFMAKDIGFENTAGTSKFQAVALR 1077
Query: 380 VGADHAVVYRCNVIGYQDTMYVHSNRQFYRECDIYGTVDFIFGNAAVVFQNCSLYARKPM 439
V AD A+ + C + G+QDT++V S RQFYR+C I GT+DF+FG+A VFQNC L R P
Sbjct: 1078VTADQAIFHNCQMDGFQDTLFVESQRQFYRDCAISGTIDFVFGDAFGVFQNCKLICRVPA 1257
Query: 440 AQQKNTITAQNRKDPNQNTGISIHACRVLAAADLKASNESFETYLGRPWKLYSRTVYMMS 499
QK +TA R N + + + L + +YLGRPWKLYS+ V M S
Sbjct: 1258KGQKCLVTAGGRDKQNSASALVFLSSHFTGEPALTSVTPKL-SYLGRPWKLYSKVVIMDS 1434
Query: 500 YIGDHVHQRGWLEWNTTFALDTLYYGEYMNYGPGAAVGQRVNWPGYRVITSALEASR--- 556
I G++ DT + EY N GPGA RV W G +V+TS + A
Sbjct: 1435TIDAMFAPEGYMPMVGGAFKDTCTFYEYNNKGPGADTNLRVKWHGVKVLTSNVAAEYYPG 1614
Query: 557 --FTVAQFILGATWLPSTGVAFLSG 579
F + TW+ +GV + G
Sbjct: 1615KFFEIVNATARDTWIVKSGVPYSLG 1689
>TC91752 similar to GP|6630559|gb|AAF19578.1| putative pectinesterase
{Arabidopsis thaliana}, partial (34%)
Length = 691
Score = 247 bits (630), Expect = 1e-65
Identities = 125/226 (55%), Positives = 163/226 (71%), Gaps = 2/226 (0%)
Frame = +3
Query: 235 RRLLAMRDDNF-PRWLNRRDRRLLSLPVSAMQ-ADVVVSKDGNGTAKTIAEAIKKVPEYG 292
RRLL++ N P WL+ +DR+LL + AD+VV+KDG+G KTI+ A+K VP
Sbjct: 18 RRLLSLSHQNEEPNWLHHKDRKLLQKDSDLKKKADIVVAKDGSGKYKTISAALKHVPNKS 197
Query: 293 SRRFIIYVRAGRYEENNLKIGRKRTNVMIIGDGKGKTVITGGKNVMDGLLTTFHTASFAA 352
+R +IYV+ G Y EN +++ + + NVMIIGDG T+++G N +DG TF TA+FA
Sbjct: 198 DKRTVIYVKKGIYYEN-VRVEKTKWNVMIIGDGMNVTIVSGSLNFIDGT-PTFSTATFAV 371
Query: 353 SGAGFIARDMTFENYAGPEKHQAVALRVGADHAVVYRCNVIGYQDTMYVHSNRQFYRECD 412
G FIARD+ F+N AGP+KHQAVAL AD AV Y+C++ YQDT+Y HSNRQFYREC+
Sbjct: 372 FGRNFIARDIGFKNTAGPQKHQAVALMTSADQAVFYKCSMDAYQDTLYAHSNRQFYRECN 551
Query: 413 IYGTVDFIFGNAAVVFQNCSLYARKPMAQQKNTITAQNRKDPNQNT 458
IYGTVDFIFGN+AVV QNC++ R+PM Q+NTITAQ + DPN NT
Sbjct: 552 IYGTVDFIFGNSAVVLQNCNILPRQPMPGQQNTITAQGKTDPNMNT 689
>TC82174 similar to GP|4097684|gb|AAD00154.1| ubiquitin conjugating enzyme
{Metarhizium anisopliae}, partial (17%)
Length = 1031
Score = 204 bits (519), Expect(2) = 3e-57
Identities = 121/209 (57%), Positives = 146/209 (68%), Gaps = 18/209 (8%)
Frame = +1
Query: 1 MEYGKLGPSDSGGGSSRSDVALVNSRTKTTSGRKKKLIILVLLAVVLIVAS---AVVLTA 57
MEYG+LG SD GG + + + TK S KKK+I L LLAV+LI+AS A +LT
Sbjct: 376 MEYGRLGLSDPGGSTPSQPII---NTTKPRSS-KKKIIFLSLLAVLLIIASTISAAMLTG 543
Query: 58 VRSRAGGSGAS-TLRAKPTQAISNTCSKTRFPSLCVKSLLDFPGSTAASERDLVHISLNM 116
+ S + TLR PTQAISNTCSKTRFPSLC+ LLDFP ST ASE+DLVHISLNM
Sbjct: 544 IHSHTTSEPKNPTLRRNPTQAISNTCSKTRFPSLCINYLLDFPDSTGASEKDLVHISLNM 723
Query: 117 TFQHLAKALYSSAAISAS--MDPRVRAAYDDCLELLDDSVDALGRSLSSFAAGSSSS--- 171
T QHL+KALY+SA+IS++ ++P +RAAY DCLELLD+SVDAL R+L+S SSS+
Sbjct: 724 TLQHLSKALYTSASISSTVGINPYIRAAYTDCLELLDNSVDALARALTSAVPSSSSNGAV 903
Query: 172 ---------DVMTWLSAALTNQDTCADGF 191
DV+TWLSAALTNQDT GF
Sbjct: 904 KPLTSSSTEDVLTWLSAALTNQDTVR*GF 990
Score = 36.6 bits (83), Expect(2) = 3e-57
Identities = 15/16 (93%), Positives = 16/16 (99%)
Frame = +2
Query: 187 CADGFADTSGDVKDQM 202
CA+GFADTSGDVKDQM
Sbjct: 977 CAEGFADTSGDVKDQM 1024
>BQ138825 similar to PIR|T49922|T49 pectin methylesterase-like protein -
Arabidopsis thaliana, partial (36%)
Length = 658
Score = 196 bits (497), Expect = 3e-50
Identities = 101/219 (46%), Positives = 137/219 (62%), Gaps = 10/219 (4%)
Frame = +1
Query: 283 EAIKKVPEYG--SRRFIIYVRAGRYEENNLKIGRKRTNVMIIGDGKGKTVITGGKNVMDG 340
EA+ P+ G +RF+IY++ G YEE +++ K+ NV+ +GDG GKTVITG NV
Sbjct: 4 EAVNAAPDNGVDRKRFVIYIKEGVYEET-VRVPLKKRNVVFLGDGIGKTVITGSANVGQP 180
Query: 341 LLTTFHTASFAASGAGFIARDMTFENYAGPEKHQAVALRVGADHAVVYRCNVIGYQDTMY 400
+TT+++A+ A G GF+A+D+T EN AGP+ HQAVA R+ +D +V+ C +G QDT+Y
Sbjct: 181 GMTTYNSATVAVLGDGFMAKDLTIENTAGPDAHQAVAFRLDSDLSVIENCEFLGNQDTLY 360
Query: 401 VHSNRQFYRECDIYGTVDFIFGNAAVVFQNCSLYAR----KPMAQQKNTITAQNRKDPNQ 456
HS RQFY+ C I G VDFIFGN+A +FQ+C + R KP + N ITA R DP Q
Sbjct: 361 AHSLRQFYKSCRIVGNVDFIFGNSAAIFQDCQILVRPRQLKPEKGENNAITAHGRTDPAQ 540
Query: 457 NTGISIHACRVLAAADLKASNES----FETYLGRPWKLY 491
+TG C + D A S + YLGRPWK Y
Sbjct: 541 STGFVFQNCLINGTEDYMALYHSNPKVHKNYLGRPWKEY 657
>BI272853 weakly similar to PIR|T04359|T04 pectin methylesterase-like protein
- maize, partial (25%)
Length = 678
Score = 191 bits (486), Expect = 5e-49
Identities = 99/208 (47%), Positives = 133/208 (63%)
Frame = +1
Query: 295 RFIIYVRAGRYEENNLKIGRKRTNVMIIGDGKGKTVITGGKNVMDGLLTTFHTASFAASG 354
R+ IYV+AG Y+E + I + N+++ GDG GKT++TG KN G+ T TA+FA +
Sbjct: 4 RYTIYVKAGVYDEY-ITIPKDAVNILMYGDGPGKTIVTGRKNGAAGV-KTMQTATFANTA 177
Query: 355 AGFIARDMTFENYAGPEKHQAVALRVGADHAVVYRCNVIGYQDTMYVHSNRQFYRECDIY 414
GFI + MTFEN AGP HQAVA R D + + C+++GYQDT+YV +NRQFYR C I
Sbjct: 178 LGFIGKAMTFENTAGPAGHQAVAFRNQGDMSALVGCHILGYQDTLYVQTNRQFYRNCVIS 357
Query: 415 GTVDFIFGNAAVVFQNCSLYARKPMAQQKNTITAQNRKDPNQNTGISIHACRVLAAADLK 474
GTVDFIFG +A + Q+ ++ R P Q NTITA NTGI I C ++ A L
Sbjct: 358 GTVDFIFGTSATLIQDSTIIVRMPSPNQFNTITADGSYVNKLNTGIVIQGCNIVPKAALF 537
Query: 475 ASNESFETYLGRPWKLYSRTVYMMSYIG 502
+ ++YLGRPWK+ ++TV M S IG
Sbjct: 538 PQRFTIKSYLGRPWKVLTKTVVMESTIG 621
>BE319906 similar to SP|Q43062|PME_ Pectinesterase PPE8B precursor (EC
3.1.1.11) (Pectin methylesterase) (PE). [Peach] {Prunus
persica}, partial (36%)
Length = 589
Score = 187 bits (476), Expect = 7e-48
Identities = 92/194 (47%), Positives = 128/194 (65%)
Frame = +2
Query: 274 GNGTAKTIAEAIKKVPEYGSRRFIIYVRAGRYEENNLKIGRKRTNVMIIGDGKGKTVITG 333
G+G + +A+ PE +R++IYV+ G Y EN ++I +K+ N+M+IG+G T+I+G
Sbjct: 2 GSGDYAKVMDAVSAAPESSMKRYVIYVKKGVYVEN-VEIKKKKWNIMLIGEGMDATIISG 178
Query: 334 GKNVMDGLLTTFHTASFAASGAGFIARDMTFENYAGPEKHQAVALRVGADHAVVYRCNVI 393
+N +DG TTF +A+FA SG GFIARD++F+N AG EKHQAVALR +D +V YRC +
Sbjct: 179 SRNYVDGS-TTFRSATFAVSGRGFIARDISFQNTAGAEKHQAVALRSDSDLSVFYRCGIF 355
Query: 394 GYQDTMYVHSNRQFYRECDIYGTVDFIFGNAAVVFQNCSLYARKPMAQQKNTITAQNRKD 453
GYQD++Y H+ RQFYREC I GTVDFIFG+A VFQNC + A++ A K K
Sbjct: 356 GYQDSLYTHTMRQFYRECKISGTVDFIFGDATAVFQNCQILAKERDA*TKEHCYCSRTKR 535
Query: 454 PNQNTGISIHACRV 467
P + C +
Sbjct: 536 PEPTNWFLLPFCNI 577
>TC84419 similar to SP|Q42920|PME_MEDSA Pectinesterase precursor (EC
3.1.1.11) (Pectin methylesterase) (PE) (P65). [Alfalfa],
partial (53%)
Length = 732
Score = 168 bits (425), Expect = 6e-42
Identities = 89/205 (43%), Positives = 118/205 (57%)
Frame = +2
Query: 353 SGAGFIARDMTFENYAGPEKHQAVALRVGADHAVVYRCNVIGYQDTMYVHSNRQFYRECD 412
+ A F A ++ FEN AG KHQAVALRV AD A+ Y C + GYQDT+Y S RQFYR+C
Sbjct: 74 TSAHFTALNVGFENSAGAAKHQAVALRVTADKALFYNCQMNGYQDTLYTQSKRQFYRDCT 253
Query: 413 IYGTVDFIFGNAAVVFQNCSLYARKPMAQQKNTITAQNRKDPNQNTGISIHACRVLAAAD 472
I GT+DF+FG+A VFQNC L RKPMA Q+ +TA R + + + C +
Sbjct: 254 ITGTIDFVFGDAVGVFQNCKLIVRKPMANQQCMVTAGGRTKVDSVSALVFQNCHFTGEPE 433
Query: 473 LKASNESFETYLGRPWKLYSRTVYMMSYIGDHVHQRGWLEWNTTFALDTLYYGEYMNYGP 532
+ YLGRPW+ +S+ V + S I G++ W +T Y EY N G
Sbjct: 434 VLTMQPKI-AYLGRPWRNFSKVVIVDSLIDGLFVPEGYMPWMGNLFKETCTYLEYNNKGA 610
Query: 533 GAAVGQRVNWPGYRVITSALEASRF 557
GAA +V WPG + I SA EA+++
Sbjct: 611 GAATNLKVKWPGVKTI-SAGEAAKY 682
>TC92372 weakly similar to GP|9759184|dbj|BAB09799.1 pectinesterase
{Arabidopsis thaliana}, partial (10%)
Length = 605
Score = 152 bits (385), Expect = 3e-37
Identities = 78/136 (57%), Positives = 105/136 (76%), Gaps = 4/136 (2%)
Frame = +3
Query: 1 MEYGKLGPSDSGGGSSRSDVALVNSRTKTTSGRK-KKLIILVLLAVVLIVASAV---VLT 56
M++ +LGPSD GG S RS L S T +S + KKLI+L +LA VLI+ASA+ ++T
Sbjct: 192 MDFDRLGPSDPGGSSRRSTAPLTPSTTSNSSPKSNKKLIMLSILAAVLIIASAISAALIT 371
Query: 57 AVRSRAGGSGASTLRAKPTQAISNTCSKTRFPSLCVKSLLDFPGSTAASERDLVHISLNM 116
VRSRA + ++ L +KPTQAIS TCSKTR+PSLC+ SLLDFPGST+ASE++LVHIS NM
Sbjct: 372 VVRSRASSNNSNLLHSKPTQAISRTCSKTRYPSLCINSLLDFPGSTSASEQELVHISFNM 551
Query: 117 TFQHLAKALYSSAAIS 132
T +H++KAL++S+ +S
Sbjct: 552 THRHISKALFASSGLS 599
>TC93392 similar to PIR|T49241|T49241 pectinesterase-like protein -
Arabidopsis thaliana, partial (18%)
Length = 467
Score = 138 bits (348), Expect = 5e-33
Identities = 62/98 (63%), Positives = 72/98 (73%)
Frame = +2
Query: 483 YLGRPWKLYSRTVYMMSYIGDHVHQRGWLEWNTTFALDTLYYGEYMNYGPGAAVGQRVNW 542
YLGRPWK YSRT++M SY+ D + GWLEWN FAL+TLYY EYMN GPGA V RV W
Sbjct: 2 YLGRPWKTYSRTIFMQSYMSDAIRPEGWLEWNGNFALNTLYYAEYMNSGPGAGVANRVKW 181
Query: 543 PGYRVITSALEASRFTVAQFILGATWLPSTGVAFLSGL 580
GY V+ + EA++FTVAQFI G LPSTGV + SGL
Sbjct: 182 SGYHVLNDSSEATKFTVAQFIEGNLGLPSTGVTYTSGL 295
>AL368659 similar to PIR|B86158|B86 hypothetical protein AAF02886.1
[imported] - Arabidopsis thaliana, partial (25%)
Length = 495
Score = 138 bits (347), Expect = 7e-33
Identities = 73/147 (49%), Positives = 98/147 (66%), Gaps = 3/147 (2%)
Frame = +1
Query: 272 KDGNGTAKTIAEAIKKVPEY---GSRRFIIYVRAGRYEENNLKIGRKRTNVMIIGDGKGK 328
+DG+G TI +AI P F+I++ G YEE + I +K+ +M+IG+G +
Sbjct: 61 QDGSGNFTTINDAIAAAPNNTVASDGYFLIFITEGVYEEY-VSIDKKKKYLMMIGEGINQ 237
Query: 329 TVITGGKNVMDGLLTTFHTASFAASGAGFIARDMTFENYAGPEKHQAVALRVGADHAVVY 388
T+ITG +NV DG TTF++A+FA GF+A ++TF N AG K+QAVALR GAD + Y
Sbjct: 238 TIITGNRNVADGF-TTFNSATFAVVAQGFVAVNITFRNTAGAAKNQAVALRSGADMSTFY 414
Query: 389 RCNVIGYQDTMYVHSNRQFYRECDIYG 415
C+ GYQDT+Y HS RQFYRECDIYG
Sbjct: 415 SCSFEGYQDTLYTHSLRQFYRECDIYG 495
>TC89457 weakly similar to SP|Q43062|PME_PRUPE Pectinesterase PPE8B
precursor (EC 3.1.1.11) (Pectin methylesterase) (PE).
[Peach] {Prunus persica}, partial (34%)
Length = 1415
Score = 132 bits (331), Expect = 5e-31
Identities = 91/277 (32%), Positives = 137/277 (48%), Gaps = 21/277 (7%)
Frame = +1
Query: 95 LLDFPGST-AASERDLVHISLNMTFQHLAKALYSSAAISASM------------DPRVRA 141
LL P ST A+S D V SL ++ A + + + R
Sbjct: 145 LLSLPHSTFASSPNDFVGSSLRVSTAKFANSAEEVITVLQKVISILSRFTNVFGHSRTSN 324
Query: 142 AYDDCLELLDDSVDALGRSLSSF--------AAGSSSSDVMTWLSAALTNQDTCADGFAD 193
A DCL+LLD S+D L +S+S+ + G + D+ TWLSA L DTC +G
Sbjct: 325 AVSDCLDLLDMSLDQLNQSISAAQKPKEKDNSTGKLNCDLRTWLSAVLVYPDTCIEGLEG 504
Query: 194 TSGDVKDQMAGNLKDLSELVSNCLAIFAGAGDDFSGVPIQNRRLLAMRDDNFPRWLNRRD 253
+ VK ++ L + LV+N L DD LA D FP W+ D
Sbjct: 505 SI--VKGLISSGLDHVMSLVANLLGEVVSGNDD----------QLATNKDRFPSWIRDED 648
Query: 254 RRLLSLPVSAMQADVVVSKDGNGTAKTIAEAIKKVPEYGSRRFIIYVRAGRYEENNLKIG 313
+LL + + AD VV+ DG+G + +A+ PE +R++IYV+ G Y E N++I
Sbjct: 649 TKLLQ--ANGVTADAVVAADGSGDYAKVMDAVSAAPESSMKRYVIYVKKGVYVE-NVEIK 819
Query: 314 RKRTNVMIIGDGKGKTVITGGKNVMDGLLTTFHTASF 350
+K+ N+M+IG+G T+I+G +N +DG TTF +A+F
Sbjct: 820 KKKWNIMLIGEGMDATIISGSRNYVDG-STTFRSATF 927
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.133 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,059,835
Number of Sequences: 36976
Number of extensions: 206849
Number of successful extensions: 1093
Number of sequences better than 10.0: 79
Number of HSP's better than 10.0 without gapping: 1005
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1014
length of query: 582
length of database: 9,014,727
effective HSP length: 101
effective length of query: 481
effective length of database: 5,280,151
effective search space: 2539752631
effective search space used: 2539752631
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0039.5


