

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0039.4
(1435 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
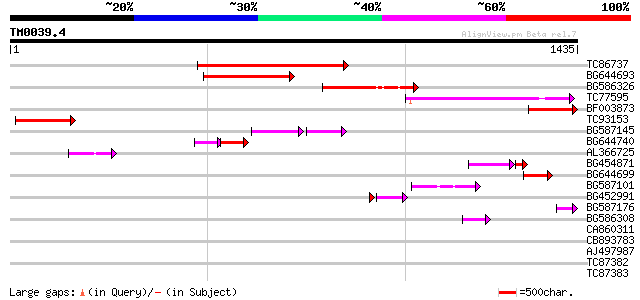
Score E
Sequences producing significant alignments: (bits) Value
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 314 1e-85
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 239 7e-63
BG586326 similar to PIR|G84493|G8 probable retroelement pol poly... 223 4e-58
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 171 2e-42
BF003873 similar to GP|14715222|em putative polyprotein {Cicer a... 151 2e-36
TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Ci... 140 4e-33
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 89 2e-29
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 87 5e-24
AL366725 85 2e-16
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 75 3e-16
BG644699 similar to PIR|T07863|T078 probable polyprotein - pinea... 66 1e-10
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 61 3e-09
BG452991 PIR|A25875|A25 histone H4 - Tetrahymena thermophila, pa... 52 1e-07
BG587176 weakly similar to PIR|G84493|G84 probable retroelement ... 47 4e-05
BG586308 weakly similar to PIR|F84528|F8 probable retroelement p... 45 3e-04
CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product ... 42 0.001
CB893783 weakly similar to GP|22830935|dbj hypothetical protein~... 42 0.002
AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis... 41 0.004
TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulch... 38 0.032
TC87383 similar to GP|19168656|emb|CAD26175. DNA-DIRECTED RNA PO... 37 0.072
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 314 bits (805), Expect = 1e-85
Identities = 175/395 (44%), Positives = 247/395 (62%), Gaps = 14/395 (3%)
Frame = +1
Query: 476 VVREFPEVF-PEDMTELPPEREV-EFAIDVIP----GTTPISAAP-YRISPLELAELQKQ 528
V+ EFP++F PE ++P R + + AI +IP P+ P Y +S EL L+K
Sbjct: 343 VLEEFPDLFNPEKAYQVPASRGLLDHAIPLIPDKDGNDPPLPWGPLYGMSRQELLVLKKT 522
Query: 529 VEELLSKGFIRPSVSPWGAPVLLVKKKDGSMRLCVDYRQLNKVTIKNRYPLPRIDDLMDQ 588
+E+LL KGFI+ S S GAPVL V+K G +R CVDYR LN +T K+RYPLP I + + +
Sbjct: 523 LEDLLDKGFIKASGSAAGAPVLFVRKPGGGIRFCVDYRALNAITKKDRYPLPLISETLRR 702
Query: 589 LKGARVFSKIDLRSGYHQIRVKSDDVQKTAFRTRYGHYEYLVMPFGVTNAPAIFMDYMNR 648
+ GAR F+K+D+ + +H++R+K +D +KTAFRTRYG +E++V PFG+T APA F Y+N+
Sbjct: 703 VAGARWFTKLDVVAAFHKMRIKDEDQEKTAFRTRYGLFEWIVCPFGLTGAPATFQRYINK 882
Query: 649 IFHPYLDKFVIVFIDDILIYSK-SKEEHVEHMQVVLKVLKDRKLYAKLSKCEFWLEQVQF 707
H +LD FV +IDD+LIY+ SK++H ++ VL+ L D L KCEF + V++
Sbjct: 883 TLHEFLDDFVTAYIDDVLIYTTGSKKDHEAQVRRVLRRLADAGLSLDPKKCEFSVTTVKY 1062
Query: 708 LGHVVSE-DGIAVDPAKVEAVNSWKVPETVTGVRSFLGLAGYYRRFIEGFSKIATPLTQL 766
+G +++ G++ DP K+ A+ W P +V G RSFLG YY+ FI G+S+I PLT+L
Sbjct: 1063VGFILTAGKGVSCDPLKLAAIRDWLPPGSVKGARSFLGFCNYYKDFIPGYSEITEPLTRL 1242
Query: 767 TKKDHPFVWTEKCE*SFQTLKERLTKAPVLTLPDPSKDYDVYCDASKSGLGCVLMQE--- 823
T+KD PF W + E +F LK + PVL + DP V D S LG VL QE
Sbjct: 1243TRKDFPFRWGAEQEAAFTKLKRLFAEEPVLRMFDPEAVTTVETDCSGFALGGVLTQEDGT 1422
Query: 824 --RKVIAYASQQLRPHEQNYPTHDMELAAVVFALK 856
+A+ SQ+L P E NYP HD EL AV L+
Sbjct: 1423GAAHPVAFHSQRLSPAEYNYPIHDKELLAVWACLR 1527
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 239 bits (609), Expect = 7e-63
Identities = 124/230 (53%), Positives = 162/230 (69%)
Frame = +2
Query: 491 LPPEREVEFAIDVIPGTTPISAAPYRISPLELAELQKQVEELLSKGFIRPSVSPWGAPVL 550
+PPE +++F ID++P PI YRI+PL+L L+ Q+++LL KGFI+PS+ P G VL
Sbjct: 17 VPPEWKIDFGIDLLPNMNPI*IPSYRINPLKLKVLKLQLKDLLEKGFIQPSIYP*GVVVL 196
Query: 551 LVKKKDGSMRLCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGARVFSKIDLRSGYHQIRVK 610
+KKKDG +R+ +DY QLN V IK +YPLP ID+L D L+G++ F KIDLR G HQ RV
Sbjct: 197 FLKKKDGFLRMSIDYPQLNNVNIKIKYPLPLIDELFDNLQGSKWFFKIDLRLG*HQHRVI 376
Query: 611 SDDVQKTAFRTRYGHYEYLVMPFGVTNAPAIFMDYMNRIFHPYLDKFVIVFIDDILIYSK 670
+DV KTAFR RYGHYE LVM FG TN P FM+ MNR+F YLD VIVF +DILIYSK
Sbjct: 377 GEDVPKTAFRIRYGHYEILVMSFG*TNPPMAFMELMNRVFQDYLDSLVIVFSNDILIYSK 556
Query: 671 SKEEHVEHMQVVLKVLKDRKLYAKLSKCEFWLEQVQFLGHVVSEDGIAVD 720
++ EH H+++ LKVLKD L ++S +E F HV+S +G+ VD
Sbjct: 557 NENEHENHLRLALKVLKDIGL-CQISYV*ILVEVGFFSLHVISGEGLKVD 703
>BG586326 similar to PIR|G84493|G8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 736
Score = 223 bits (568), Expect = 4e-58
Identities = 122/247 (49%), Positives = 157/247 (63%), Gaps = 2/247 (0%)
Frame = +2
Query: 791 TKAPVLTLPDPSKDYDVYCDASKSGLGCVLMQERKVIAYASQQLRPHEQNYPTHDMELAA 850
T AP+L LP+ Y VY DAS +GLGCVL Q KVIAYAS+QLR HE NYPTHD+E+AA
Sbjct: 8 TSAPILVLPELIT-YVVYTDASITGLGCVLTQHEKVIAYASRQLRKHEGNYPTHDLEMAA 184
Query: 851 VVFALKIWRHYLYGVKFTIYSDHQSLKYLFDQKTLNMRQRRWVEFLEDYDFKLQYHPGKA 910
VVFALKIWR YLYG K I++DH+SLKY+F Q LN+RQRRW+EF+ DYD + Y+PGKA
Sbjct: 185 VVFALKIWRSYLYGAKVQIHTDHKSLKYIFTQPELNLRQRRWMEFVADYDLDITYYPGKA 364
Query: 911 NVVADALSRKSLHAARLMIEETELIEKFRDMNLIMETLPQGTRLGTLTLTN--EFIEEVK 968
N+VADALSR+ + + E +L R + L L + T L N + ++
Sbjct: 365 NLVADALSRRRVDVSAER-EADDLDGMVRALRL--NVLTKATESLGLEAVNQADLFTRIR 535
Query: 969 KEQARDENLQKEAHGRDSMSRPDFLKGPDGLWRYQGRLCVPEGGELRQKILEEGHKSDFS 1028
Q +DENLQK A R ++ DG GR+ VP L+++I+ E HKS FS
Sbjct: 536 LAQGQDENLQKVAQN----DRTEYQTAKDGTILVNGRISVPNDRSLKEEIMSEAHKSRFS 703
Query: 1029 IHPGTTK 1035
+HPG +
Sbjct: 704 VHPGAPR 724
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial (14%)
Length = 1708
Score = 171 bits (432), Expect = 2e-42
Identities = 126/451 (27%), Positives = 213/451 (46%), Gaps = 23/451 (5%)
Frame = +2
Query: 1002 YQGRLCVPEG-------GELRQKILEEGHKSDFSIHPGTTKMYQDLKKMFWWPGMKKDIM 1054
++GR+ VP ELR K+++E H S + HPG + + + F+WPG + +
Sbjct: 110 FRGRIWVPGSDDEESPLNELRTKLVQESHDSTAAGHPGRNGTLEIVSRKFFWPGQSQTVR 289
Query: 1055 KKVTSCLTCQKVKGEHQKPSGSLQPLSIPEWKWEGISMDFVSGLPRTT-TGHDAIWVIVD 1113
+ V +C C + Q G L+PL +P +SMDF++ LP T G +WVIVD
Sbjct: 290 RFVRNCDVCGGIHIWRQAKRGFLKPLPVPNRLHSDLSMDFITSLPPTRGRGSQYLWVIVD 469
Query: 1114 RLTKSAHFIAVNMTFPSEKLARIYVKEIVRLHGVPANIVSDRDPRFVSKFWGSLHEALGT 1173
RL+KS ++ T +E A+ ++ R HG+P +IVSDR +V +FW G
Sbjct: 470 RLSKSVTLEEMD-TMEAEACAQRFLSCHYRFHGMPQSIVSDRGSNWVGRFWREFCRLTGV 646
Query: 1174 RLSLSSAYHPQSDGQSERTIQTLEDMLRACVLDYKGSWEDFLPLAEFSYNNSYHSSLGMA 1233
LS++YHPQ+DG +ER Q ++ +LRA V + +W D LP + + N ++SS+G
Sbjct: 647 TQLLSTSYHPQTDGGTERWNQEIQAVLRAYVCWSQDNWGDLLPTVQLALRNRHNSSIGAT 826
Query: 1234 PFEALYGRRCKTPLCWLSGEDKITLGPE-----LLQEMTEKVRSIREKLRIAQDRQKSYY 1288
PF +G P+ + + E L++ M + I+ ++ AQ R ++
Sbjct: 827 PFFVEHGYHV-DPIPTVEDTGGVVSEGEAAAQLLVKRMKDVTGFIQAEIVAAQQRSEASA 1003
Query: 1289 DKRHKPLE-FQEGDHVFLRVTPITGVGRS-----IHSKKLTPKYLGPYQILDRIGAVAYR 1342
+KR P + +Q GD V+L V+ S +H K +++ P+ + + Y
Sbjct: 1004 NKRRCPADRYQVGDKVWLNVSNYKSPRPSKKLDWLHHKYEVTRFVTPHVVELNVPGTVY- 1180
Query: 1343 IALPPSLSNLHDVFHISQLRKYLPD---SSHVIEPDNIEL-EENLTYPTQPVKILERREK 1398
FH+ LR+ D V++P + +++ + +IL R
Sbjct: 1181 -----------PKFHVDLLRRAASDPLPGQEVVDPQPPPIVDDDGEVEWEVEEILAARWH 1327
Query: 1399 QLRKRTVPLVKLAWSDDNQDATWELEESARK 1429
Q+ + + W DATWE ++ R+
Sbjct: 1328 QVGRGRRRQALVKWK-GFVDATWEAADAIRE 1417
>BF003873 similar to GP|14715222|em putative polyprotein {Cicer arietinum},
partial (82%)
Length = 559
Score = 151 bits (381), Expect = 2e-36
Identities = 69/125 (55%), Positives = 94/125 (75%), Gaps = 1/125 (0%)
Frame = +2
Query: 1312 GVGRSIHSKKLTPKYLGPYQILDRIGAVAYRIALPPSLSNLHDVFHISQLRKYLPDSSHV 1371
GVGR++ SKKLT +++GPYQI +R+G VAYR+ LPP L NLHDVFH+SQLRKY+PD SHV
Sbjct: 2 GVGRALKSKKLTVRFIGPYQISERVGTVAYRVGLPPHLLNLHDVFHVSQLRKYVPDPSHV 181
Query: 1372 IEPDNIELEENLTYPTQPVKILERREKQLRKRTVPLVKLAWSDDN-QDATWELEESARKR 1430
I+ D++++ +NLT T PV+I +R+ K LR + +PLV++ W N + TWELE +
Sbjct: 182 IQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVVWDRANGESLTWELESKMVES 361
Query: 1431 YPSLF 1435
YP LF
Sbjct: 362 YPELF 376
>TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Cicer
arietinum}, partial (8%)
Length = 516
Score = 140 bits (353), Expect = 4e-33
Identities = 65/155 (41%), Positives = 102/155 (64%), Gaps = 2/155 (1%)
Frame = +2
Query: 14 ARAEQQNQPAE--DDVYKGIDKFLKRNPPLFDGGYDPEGANRWLRKIEQIYESLPTSEDR 71
A+A QQ + D + ++ FL+ +PP F G Y P+GA +WL++IE+I+ + E +
Sbjct: 50 AQAVQQLPKVDTGSDGTRMLETFLRNHPPTFKGRYAPDGA*KWLKEIERIFRVMQCFETQ 229
Query: 72 MIAYASYLFHEEARNWWVHAKSRITPPDGVLTWSIFKEAFLEKYFPADVKGKKETEFLEL 131
+ + +++ EEA +WW+ + D V+TW++F++ FL +YFP DV+GKKE EFLEL
Sbjct: 230 KVQFGTHMLAEEADDWWISLLPVLEQDDAVVTWAMFRKEFLGRYFPEDVRGKKEIEFLEL 409
Query: 132 KQGEMFVGQYAARFEELSQFHPYYGTTADDASKCI 166
KQG+M V +YAA+F EL+ F+P+Y + SKCI
Sbjct: 410 KQGDMSVTEYAAKFVELATFYPHYSAETAEFSKCI 514
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 88.6 bits (218), Expect(2) = 2e-29
Identities = 46/133 (34%), Positives = 76/133 (56%)
Frame = +2
Query: 612 DDVQKTAFRTRYGHYEYLVMPFGVTNAPAIFMDYMNRIFHPYLDKFVIVFIDDILIYSKS 671
DD++KTAF T G Y Y VMPFG+ NA + + +NR+F L + V+IDD+L+ S
Sbjct: 11 DDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVKSLR 190
Query: 672 KEEHVEHMQVVLKVLKDRKLYAKLSKCEFWLEQVQFLGHVVSEDGIAVDPAKVEAVNSWK 731
+H+ H++ K L + + +KC F + +FLG++V++ GI V+P ++ A+
Sbjct: 191 ATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQGIEVNPKQITAILDLP 370
Query: 732 VPETVTGVRSFLG 744
P+ V+ G
Sbjct: 371 SPKNSREVQRLTG 409
Score = 60.5 bits (145), Expect(2) = 2e-29
Identities = 36/106 (33%), Positives = 55/106 (50%), Gaps = 4/106 (3%)
Frame = +3
Query: 751 RFIEGFSKIATPLTQLTKKDHPFVWTEKCE*SFQTLKERLTKAPVLTLPDPSKDYDVYCD 810
RFI + P +L + FVW EKCE +F+ LK+ LT PVL+ P+ +Y
Sbjct: 429 RFISRSTDKCLPFYKLLCGNKRFVWDEKCEEAFEQLKQYLTTPPVLSKPEAGDTLSLYIA 608
Query: 811 ASKSGLGCVLMQ----ERKVIAYASQQLRPHEQNYPTHDMELAAVV 852
S + + VL++ E+K I Y S+++ E YPT + AV+
Sbjct: 609 ISSTAVSSVLIREDRGEQKPIFYTSKRMTDPETRYPTLEKMAFAVI 746
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 87.0 bits (214), Expect(2) = 5e-24
Identities = 41/69 (59%), Positives = 52/69 (74%)
Frame = -1
Query: 535 KGFIRPSVSPWGAPVLLVKKKDGSMRLCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGARV 594
K F +PS+SP GA +L V+KKDG R+C+DYRQ NKVT KN+YPLPRID+L D+++
Sbjct: 274 KRFQQPSISP*GAALLFVRKKDGYFRMCIDYRQFNKVTTKNKYPLPRIDNLFDKIQEDCY 95
Query: 595 FSKIDLRSG 603
F IDLR G
Sbjct: 94 F*NIDLRLG 68
Score = 43.9 bits (102), Expect(2) = 5e-24
Identities = 25/69 (36%), Positives = 40/69 (57%)
Frame = -2
Query: 469 PAMEDIPVVREFPEVFPEDMTELPPEREVEFAIDVIPGTTPISAAPYRISPLELAELQKQ 528
P E + V++ F VFP++ +P ERE+ F ID++ T IS P + EL +L+
Sbjct: 471 PLFEVVLVLKGFS*VFPDNFPVIPLEREIFFCIDLLLDTQLISNPP*LMDRTELKKLKI* 292
Query: 529 VEELLSKGF 537
+++ L KGF
Sbjct: 291 LKDSLEKGF 265
>AL366725
Length = 485
Score = 85.1 bits (209), Expect = 2e-16
Identities = 45/120 (37%), Positives = 69/120 (57%)
Frame = +2
Query: 150 QFHPYYGTTADDASKCIRFECGLRPDIRAAIGHQQIRTFTVLVEKCRIFEENDRARREYY 209
+F+P+Y + SKCI+FE GLRPDI+ AIG+QQ+R F LV CRI+EE+ +A +
Sbjct: 2 KFYPHYAAETAEFSKCIKFENGLRPDIKRAIGYQQLRVFPDLVNTCRIYEEDTKAHDKVV 181
Query: 210 KSSKFNKTSKRREERKKPYSPRNYKPELQNRNYGGARPTNPNSHVTCYRCGKEGHKSWSC 269
K +K + R KPYS K + + + + + + + C+ G++GHKS C
Sbjct: 182 NERK----TKGQ*SRPKPYSAPADKGKQRMVDDRRPKKKDAPAEIVCFNYGEKGHKSNVC 349
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 75.1 bits (183), Expect(2) = 3e-16
Identities = 48/120 (40%), Positives = 64/120 (53%), Gaps = 3/120 (2%)
Frame = +2
Query: 1161 SKFWGSLHEALGTRLSLSSAYHPQSDGQSERTIQTLEDMLRACVLDYKGSWEDFLPLAEF 1220
S FW L + GT L++SSAYHP SDGQSE + E LR + W P AE+
Sbjct: 32 SNFWKQLFKLHGTILTMSSAYHP*SDGQSEALNKGXEMYLRCLMFTDPLKWSKAFPWAEY 211
Query: 1221 SYNNSYHSSLGMAPFEALYGRRCKTPL-CWLSGEDKITLGPELLQ--EMTEKVRSIREKL 1277
YN SY+ S M PF+ALYGR + S +D L +L Q E+ +++SI +L
Sbjct: 212 WYNTSYNISAAMTPFKALYGRDLSMLIRSKGSSKDTADLQSQLAQREELLSQLQSISTRL 391
Score = 29.6 bits (65), Expect(2) = 3e-16
Identities = 12/30 (40%), Positives = 18/30 (60%)
Frame = +1
Query: 1280 AQDRQKSYYDKRHKPLEFQEGDHVFLRVTP 1309
AQ K DK+ + EFQ G+HV +++ P
Sbjct: 388 AQQTMKHQADKKRRHFEFQLGEHVLVKLQP 477
>BG644699 similar to PIR|T07863|T078 probable polyprotein - pineapple
retrotransposon dea1 (fragment), partial (5%)
Length = 231
Score = 65.9 bits (159), Expect = 1e-10
Identities = 31/73 (42%), Positives = 48/73 (65%), Gaps = 1/73 (1%)
Frame = +2
Query: 1301 DHVFLRVTPIT-GVGRSIHSKKLTPKYLGPYQILDRIGAVAYRIALPPSLSNLHDVFHIS 1359
+ V L+V P G R KL+ +Y+GP++++ RIG VAY +ALPP LS +H VFH+S
Sbjct: 2 EQVLLKVLPTERGDCRFGKRGKLSLRYIGPFEVIKRIGEVAYELALPPGLSGVHPVFHVS 181
Query: 1360 QLRKYLPDSSHVI 1372
++Y D +++I
Sbjct: 182 MFKRYHGDGNYII 220
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 61.2 bits (147), Expect = 3e-09
Identities = 45/177 (25%), Positives = 87/177 (48%), Gaps = 4/177 (2%)
Frame = +2
Query: 1018 ILEEGHKSDFSIHPGTTKMYQDLKKM-FWWPGMKKDIMKKVTSCLTCQK---VKGEHQKP 1073
IL H S+++ H +K +++ FWWP M KD ++ C CQ+ + ++ P
Sbjct: 104 ILFHCHGSNYAGHFAVSKTVSKIQQAGFWWPTMFKDAHSFISKCDPCQRQGNIS*RNEMP 283
Query: 1074 SGSLQPLSIPEWKWEGISMDFVSGLPRTTTGHDAIWVIVDRLTKSAHFIAVNMTFPSEKL 1133
+ + + ++ +DF+ P ++ + I V VD ++K IA + T + +
Sbjct: 284 QNFILEVEV----FDVWGIDFMGPFP-SSYNNKYILVAVDYVSKWVEAIA-SPTNDATVV 445
Query: 1134 ARIYVKEIVRLHGVPANIVSDRDPRFVSKFWGSLHEALGTRLSLSSAYHPQSDGQSE 1190
+++ I GVP ++SD F++K + L + G R +++AYHPQ +S+
Sbjct: 446 VKMFKSVIFPRFGVPRVVISDGGSHFINKVFEKLLKKNGVRHKVATAYHPQKAERSK 616
>BG452991 PIR|A25875|A25 histone H4 - Tetrahymena thermophila, partial (33%)
Length = 560
Score = 52.0 bits (123), Expect(2) = 1e-07
Identities = 24/78 (30%), Positives = 42/78 (53%)
Frame = +3
Query: 929 IEETELIEKFRDMNLIMETLPQGTRLGTLTLTNEFIEEVKKEQARDENLQKEAHGRDSMS 988
+E +E+FRD++L+ E PQ +LG L + NEF++ +K+ Q D L G +
Sbjct: 51 LESWSCLEQFRDLSLVCEVSPQSVKLGMLKINNEFLDSIKEAQKVDVKLVDLMFGNNQTE 230
Query: 989 RPDFLKGPDGLWRYQGRL 1006
DF G+ +++ R+
Sbjct: 231 DGDFKVDDQGVLQFRDRI 284
Score = 23.5 bits (49), Expect(2) = 1e-07
Identities = 10/12 (83%), Positives = 11/12 (91%)
Frame = +1
Query: 912 VVADALSRKSLH 923
VVAD LSRK+LH
Sbjct: 1 VVADVLSRKTLH 36
>BG587176 weakly similar to PIR|G84493|G84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(1%)
Length = 729
Score = 47.4 bits (111), Expect = 4e-05
Identities = 21/54 (38%), Positives = 32/54 (58%), Gaps = 1/54 (1%)
Frame = -1
Query: 1383 LTYPTQPVKILERREKQLRKRTVPLVKLAWS-DDNQDATWELEESARKRYPSLF 1435
L T+PV+IL+R EK +RK+ + +VK+ W ++ TWE E + YP F
Sbjct: 717 LDLETRPVRILDRMEKAMRKKPIQMVKIVWDCSGREEITWETEARMKADYPEWF 556
>BG586308 weakly similar to PIR|F84528|F8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 686
Score = 44.7 bits (104), Expect = 3e-04
Identities = 25/71 (35%), Positives = 39/71 (54%)
Frame = -2
Query: 1145 HGVPANIVSDRDPRFVSKFWGSLHEALGTRLSLSSAYHPQSDGQSERTIQTLEDMLRACV 1204
HG+P IV+D F+S + E RL+ +S +PQS+GQ+E + + + D L+ +
Sbjct: 685 HGLPYEIVTDNGSHFISNKFREFCERWRIRLNTASPRYPQSNGQAEASNKIIIDGLKKRL 506
Query: 1205 LDYKGSWEDFL 1215
KG W D L
Sbjct: 505 DLKKGCWADEL 473
>CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product {Drosophila
melanogaster}, partial (20%)
Length = 192
Score = 42.4 bits (98), Expect = 0.001
Identities = 23/60 (38%), Positives = 36/60 (59%), Gaps = 5/60 (8%)
Frame = +1
Query: 815 GLGCVLMQERKV-----IAYASQQLRPHEQNYPTHDMELAAVVFALKIWRHYLYGVKFTI 869
G+G L Q+ + IAYAS+ L E+NY + E A ++A++ +RHYL+G KF +
Sbjct: 13 GIGAGLSQKDEENHEHPIAYASRLLTAAERNYTVVERECLAAIWAIRNFRHYLHGPKFEL 192
>CB893783 weakly similar to GP|22830935|dbj hypothetical protein~similar to
gag-pol polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 853
Score = 41.6 bits (96), Expect = 0.002
Identities = 49/205 (23%), Positives = 83/205 (39%), Gaps = 33/205 (16%)
Frame = +2
Query: 335 LCDISLVVLYDSGATHSFISHERAKSLKLVITQLPYDLVVTTPTKESAVTSSVCKKCPLV 394
+CD V+ DSG+ + +S+ + L+L P+ + K + V S C
Sbjct: 257 VCD----VIIDSGSCENVVSNYMVEKLELPTKDHPHRYKLQWLKKGNEVRVSKCCLVSFS 424
Query: 395 IEDREYITNLVC--LPLEGLDIILGMNWLSINNVLLDCRLRVPIFLQ------------- 439
I ++Y N+ C + ++ ++LG W + L D F++
Sbjct: 425 I-GQKYKDNVWCDVISMDACHMLLGRPWQYDRHALYDGHANTYTFVKYGVKIKLVPLPPN 601
Query: 440 -----KYKEKHTASLPEKEPSAY------------LILFSSEGTKRPAMEDIPVVREFPE 482
K K SL KEP L+ + E T + +E + V +F +
Sbjct: 602 AFDEGKKDFKPIVSLVSKEPFKVTTKDIQDMSLILLVKSNEESTIQKEVEHLLV--DFTD 775
Query: 483 VFPEDMTE-LPPEREVEFAIDVIPG 506
V P ++ LPP R+++ AID IPG
Sbjct: 776 VVPSEIPSGLPPMRDIQHAIDFIPG 850
>AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis thaliana
chromosome II BAC F26H6; putative retroelement pol
polyprotein, partial (1%)
Length = 636
Score = 40.8 bits (94), Expect = 0.004
Identities = 23/78 (29%), Positives = 39/78 (49%), Gaps = 1/78 (1%)
Frame = -2
Query: 850 AVVFALKIWRHYLYGVKFTIYSDHQSLKYLFDQKTLNMRQRRWVEFLEDYDFKLQYHPG- 908
A+ +A K RHY+ + S +KY+F++ L R RW L +YD + +
Sbjct: 623 ALAWAAKRLRHYMINHTTWLVSKMDPIKYIFEKPALTGRIARWQMLLSEYDIEYRSQKAI 444
Query: 909 KANVVADALSRKSLHAAR 926
K +++AD L+ + L R
Sbjct: 443 KGSILADHLAHQPLEDYR 390
>TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulchellus},
partial (7%)
Length = 2304
Score = 37.7 bits (86), Expect = 0.032
Identities = 44/192 (22%), Positives = 80/192 (40%), Gaps = 15/192 (7%)
Frame = +2
Query: 35 LKRNPPLFDGGYDPEGANRWLRKIEQIYESLPTSEDRMIAYASYLFHEEARNWWVHAKSR 94
+K + P F+G + WL+ IE+++E E++ + + + A WW + K R
Sbjct: 614 IKVDIPDFEGNLQLDDFLDWLQTIERVFEYKEVPEEQKVKIVAAKLKKHALIWWENLKRR 793
Query: 95 --ITPPDGVLTWSIFKEAFLEKYFP-----ADVKGK---KETEFLELKQGEMFVGQYAAR 144
+ TW ++ KY P A+ K K++ + L + + +
Sbjct: 794 RKREGKSKIKTWDKMRQKLTRKYLPPHYYQANFTQK*LPKKSSYQPLSPTKNHIDYHKPL 973
Query: 145 FEE-LSQFHPYYGTTAD---DASKCIRFECGLRPDIRAAIGHQQIRTFTVLVEKC-RIFE 199
+ +S F P T + + KC F C I +Q++ FT++ E+ IFE
Sbjct: 974 IHQPISSFRPQRNTIKERNTNIPKC--FICQGYGHIALDCVNQKV--FTIVNEEINNIFE 1141
Query: 200 ENDRARREYYKS 211
E R + Y+S
Sbjct: 1142EE---REDVYES 1168
>TC87383 similar to GP|19168656|emb|CAD26175. DNA-DIRECTED RNA POLYMERASE II
{Encephalitozoon cuniculi}, partial (0%)
Length = 1247
Score = 36.6 bits (83), Expect = 0.072
Identities = 19/85 (22%), Positives = 38/85 (44%), Gaps = 2/85 (2%)
Frame = -2
Query: 35 LKRNPPLFDGGYDPEGANRWLRKIEQIYESLPTSEDRMIAYASYLFHEEARNWWVHAKSR 94
+K + P F+G P+ WL+ +E++++ E++ + + + A WW + K R
Sbjct: 739 IK*DIPDFEGNLQPDDLLDWLQIMERLFKYKEVLEEQKVKIVAAKLKKLASIWWENVKRR 560
Query: 95 --ITPPDGVLTWSIFKEAFLEKYFP 117
+ TW ++ KY P
Sbjct: 559 RKREGKSKIKTWEKMRQKLTRKYLP 485
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.136 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 44,440,311
Number of Sequences: 36976
Number of extensions: 630017
Number of successful extensions: 3134
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 3054
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3124
length of query: 1435
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1327
effective length of database: 5,021,319
effective search space: 6663290313
effective search space used: 6663290313
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0039.4


