

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0037a.4
(616 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
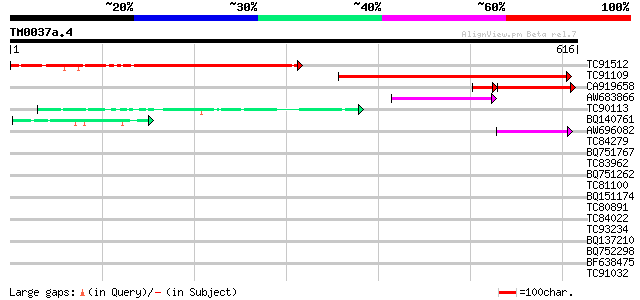
Score E
Sequences producing significant alignments: (bits) Value
TC91512 similar to PIR|D85359|D85359 hypothetical protein AT4g30... 370 e-102
TC91109 similar to GP|15028145|gb|AAK76696.1 unknown protein {Ar... 219 2e-57
CA919658 weakly similar to PIR|D85359|D853 hypothetical protein ... 117 3e-34
AW683866 similar to GP|15217302|gb| Hypothetical protein {Oryza ... 75 7e-14
TC90113 similar to PIR|C86301|C86301 arginine/serine-rich protei... 48 1e-05
BQ140761 weakly similar to GP|18076245|emb phosphophoryn {Rattus... 43 3e-04
AW696082 42 7e-04
TC84279 weakly similar to GP|1799947|dbj|BAA16433.1 BENZENE 1 2-... 42 0.001
BQ751767 similar to SP|O10302|GTA_N Probable global transactivat... 40 0.002
TC83962 similar to PIR|B86289|B86289 hypothetical protein AAF719... 40 0.002
BQ751262 similar to GP|6715100|gb|A versicolorin B synthase {Asp... 40 0.003
TC81100 similar to PIR|H84824|H84824 En/Spm-like transposon prot... 40 0.003
BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xy... 40 0.003
TC80891 similar to GP|21622328|emb|CAD36969. probable isoamyl al... 39 0.004
TC84022 homologue to GP|15822513|gb|AAG23338.1 thiamine biosynth... 39 0.006
TC93234 weakly similar to GP|21751020|dbj|BAC03887. unnamed prot... 38 0.010
BQ137210 similar to SP|P09789|GRP1_ Glycine-rich cell wall struc... 38 0.010
BQ752298 similar to GP|11595639|em conserved hypothetical protei... 38 0.013
BF638475 similar to GP|9502414|gb| peroxisomal targeting signal ... 38 0.013
TC91032 weakly similar to GP|20196995|gb|AAB91977.2 expressed pr... 37 0.017
>TC91512 similar to PIR|D85359|D85359 hypothetical protein AT4g30710
[imported] - Arabidopsis thaliana, partial (23%)
Length = 1285
Score = 370 bits (949), Expect = e-102
Identities = 222/336 (66%), Positives = 251/336 (74%), Gaps = 18/336 (5%)
Frame = +2
Query: 1 MDVCESQQASSRRLKAVETPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPS-- 58
MDVCES+QA R+ + ETPR PLV AEKNNA RRS TREV+SRYKSPTP +
Sbjct: 290 MDVCESEQAL-RKHRTRETPRRPLVPAEKNNAIPT----RRSGTREVSSRYKSPTPAAAS 454
Query: 59 ------TPSGSRRCPSPNLTR--------TATPAASLQLLPKRSLSAERKRPSTPPSPPS 104
+PSG+ RCPSP+ R T T AAS +LLPKR+LSAERKRP+TP SPPS
Sbjct: 455 GPQRCPSPSGTHRCPSPSGPRRCPSPSSFTRTTAAS-KLLPKRALSAERKRPTTPTSPPS 631
Query: 105 RRLSTPVNDSAIDGRLSSRKVAASRLPEGHNLWPSTMRSLSVSFQSDIISIPVSKKERPV 164
STP D D LS R+ A+SR+ E +LWPSTMRSLSVSFQSD ISIPVSKKERPV
Sbjct: 632 P--STPAQD---DVHLS-RRAASSRMQE--SLWPSTMRSLSVSFQSDTISIPVSKKERPV 787
Query: 165 TSASDRTLRPASNVAHRQGETPSTIRKATPERKRSPLKGKNASDQSENSKPVDSLPSRLI 224
TSASDRTLRP SNVAH+Q ETP+T RK TPER+RSPLKGKNASDQSENSKPVD L SRLI
Sbjct: 788 TSASDRTLRPTSNVAHKQVETPNT-RKPTPERRRSPLKGKNASDQSENSKPVDGLQSRLI 964
Query: 225 DQHRWPSRIGGKVSSSALNRSVDFGD--ARMLKTPASGTGFSSLRRLSLSEEASKPLQRA 282
DQHRWPSRIGGKVSS++LNRSVD D R L + G+G SSLRR SL +AS+PLQ+
Sbjct: 965 DQHRWPSRIGGKVSSNSLNRSVDHSDKITRTLNSSVPGSGVSSLRRFSLPGDASRPLQKT 1144
Query: 283 SSDSVRLLSLVASGRTGSEVKSVDDCSLHDLRPHRS 318
S+D RLLSLV +GR GSEVK+ DD S LRPH+S
Sbjct: 1145STDVARLLSLVENGRIGSEVKAFDD-SFQVLRPHKS 1249
Score = 36.2 bits (82), Expect = 0.038
Identities = 60/248 (24%), Positives = 91/248 (36%), Gaps = 31/248 (12%)
Frame = +2
Query: 180 HRQGETPSTIRKATPERKRSPLKGKNASDQSENSKPVDSLPSRLIDQHRWPSRIGGKVSS 239
HR ETP R P K + + + + + +S+ P+ R PS
Sbjct: 326 HRTRETPR--RPLVPAEKNNAIPTRRSGTREVSSRYKSPTPAAASGPQRCPS-------P 478
Query: 240 SALNRSVDFGDARMLKTPASGTGFSSLRRL----SLSEEASKPLQRAS--------SDSV 287
S +R R +P+S T ++ +L +LS E +P S D V
Sbjct: 479 SGTHRCPSPSGPRRCPSPSSFTRTTAASKLLPKRALSAERKRPTTPTSPPSPSTPAQDDV 658
Query: 288 RLLSLVASGRTG-----SEVKSV------DDCSLHDLRPHRSSTPHTDRAGLSIAGVRSQ 336
L AS R S ++S+ D S+ + R T +DR + V +
Sbjct: 659 HLSRRAASSRMQESLWPSTMRSLSVSFQSDTISIPVSKKERPVTSASDRTLRPTSNVAHK 838
Query: 337 SLSAPGSRLPSPSRISVQSTSSSRGVSPSRSRPSTPPSRGGVS--------PSRIRPTSS 388
+ P +R P+P R S +G + S ++ P G S PSRI S
Sbjct: 839 QVETPNTRKPTPER----RRSPLKGKNASDQSENSKPVDGLQSRLIDQHRWPSRIGGKVS 1006
Query: 389 SIQSNDSV 396
S N SV
Sbjct: 1007SNSLNRSV 1030
>TC91109 similar to GP|15028145|gb|AAK76696.1 unknown protein {Arabidopsis
thaliana}, partial (35%)
Length = 1166
Score = 219 bits (558), Expect = 2e-57
Identities = 121/258 (46%), Positives = 173/258 (66%), Gaps = 5/258 (1%)
Frame = +1
Query: 358 SSRGVSPSR--SRPSTPPSRGGVSPSRIRPT-SSSIQSND--SVSVLSFIADFRKGKKGA 412
++R SPS+ + ++ PSRG SPS++R +S+I SN S S+LSF AD R+GK G
Sbjct: 19 NARPASPSKLWASAASSPSRGFSSPSKVRSAVASTINSNSGSSPSILSFSADVRRGKIGE 198
Query: 413 AFIEDAHQLRLLYNRFMQWRFANARAEAVLYIQNAIVQKTLYNVWIATLSLWESVTRKKI 472
I DAH LRLLYNR++QWRF NARA+A +Q + L+N W+ L SV K+I
Sbjct: 199 DRIFDAHTLRLLYNRYVQWRFVNARADAAFMVQKLNAETHLWNAWVTISELRHSVILKRI 378
Query: 473 NLQQLKLELKLNSVLNDQMAYLDEWATLETDHVDALSGAVEDLEASTLRLPVTRGAMVDI 532
L L+ +LKL S+L Q++YL+EWA L+ DH ++ GA E L ASTLRLP+ A D+
Sbjct: 379 KLVLLRQKLKLTSILKGQISYLEEWALLDRDHSSSVLGATEALRASTLRLPLVEKATADV 558
Query: 533 EHLKVAICQAVDVMQAMGSAIRSLFSQVEGMNDLISGVAVVATKEKSILDECEMLLASVA 592
+LK A+ AVDVMQAM S+I SL S+VE N L++ + V +KE+ +L +C+ L+S+A
Sbjct: 559 PNLKDALGSAVDVMQAMASSIYSLSSKVEETNCLVAEILKVTSKERFLLQQCKDFLSSLA 738
Query: 593 ALQVEESSLQTHLIQFKQ 610
A+QV++ SL+TH++Q +
Sbjct: 739 AMQVKDCSLKTHMLQLSR 792
>CA919658 weakly similar to PIR|D85359|D853 hypothetical protein AT4g30710
[imported] - Arabidopsis thaliana, partial (13%)
Length = 809
Score = 117 bits (292), Expect(2) = 3e-34
Identities = 60/84 (71%), Positives = 72/84 (85%)
Frame = -3
Query: 531 DIEHLKVAICQAVDVMQAMGSAIRSLFSQVEGMNDLISGVAVVATKEKSILDECEMLLAS 590
DIE LKVAIC AVDVMQAMGSAIR LFS+VEGMN+LIS VA+V+ +EK++LDECE LLA
Sbjct: 732 DIEPLKVAICSAVDVMQAMGSAIRPLFSRVEGMNNLISEVAIVSAQEKAMLDECEALLAF 553
Query: 591 VAALQVEESSLQTHLIQFKQVLGI 614
++QVEE SL+THL+QFKQ L +
Sbjct: 552 STSMQVEEYSLRTHLMQFKQALEV 481
Score = 47.0 bits (110), Expect(2) = 3e-34
Identities = 22/27 (81%), Positives = 25/27 (92%)
Frame = -1
Query: 504 HVDALSGAVEDLEASTLRLPVTRGAMV 530
HVDALSGAVEDLEA+TLRLP+T GA +
Sbjct: 809 HVDALSGAVEDLEANTLRLPLTGGAKI 729
>AW683866 similar to GP|15217302|gb| Hypothetical protein {Oryza sativa}
[Oryza sativa (japonica cultivar-group)], partial (4%)
Length = 552
Score = 75.1 bits (183), Expect = 7e-14
Identities = 40/113 (35%), Positives = 60/113 (52%)
Frame = +3
Query: 416 EDAHQLRLLYNRFMQWRFANARAEAVLYIQNAIVQKTLYNVWIATLSLWESVTRKKINLQ 475
ED H+LRLL NR +QWR+ANA A V + + L VW +V +KKI
Sbjct: 93 EDVHKLRLLDNRLIQWRYANAXAHIVNANISRHTESNLICVWDGLTKSRNTVMKKKIQFA 272
Query: 476 QLKLELKLNSVLNDQMAYLDEWATLETDHVDALSGAVEDLEASTLRLPVTRGA 528
+ KL +K +L Q+ L+ W ++E HV ++ E L ++ R+P+ GA
Sbjct: 273 REKLXMKKAFILYYQLKLLEAWGSMERQHVSTITATKECLHSAVCRIPLLEGA 431
>TC90113 similar to PIR|C86301|C86301 arginine/serine-rich protein
[imported] - Arabidopsis thaliana, partial (70%)
Length = 1259
Score = 47.8 bits (112), Expect = 1e-05
Identities = 87/362 (24%), Positives = 134/362 (36%), Gaps = 8/362 (2%)
Frame = +1
Query: 31 NAATATATARRSRTREVTSRYKSPTPPSTPSGSRRCPSPNLTRTATPAASLQLLPKRSLS 90
++++ +++ RSR+R +S +SP+ S+ S SR P P +A PA + P
Sbjct: 31 SSSSRSSSRSRSRSRSFSSSSRSPSR-SSKSRSRSPPPPRRKSSAEPARRGRS-PPLPPP 204
Query: 91 AERKRPSTPPSPPSRRLSTPVNDSAIDGRLSSRKVAASRLPEGHNLWPSTMRSLSVSFQS 150
+ KRPS PP PS PV +S + L K++ + + EGH ++ + +F
Sbjct: 205 PQSKRPSPPPRKPS-----PVRESLV---LHVEKLSRN-VNEGH------LKEIFSNF-G 336
Query: 151 DIISIPVSKKERPVTSASDRTLRPASNVAHRQGETPSTIRKATPERKRSPLKGKNAS--- 207
+++S+ + DR + + +T KA + + G
Sbjct: 337 EVVSVEL---------VMDRAVNLPKGYGYVHFKTRGEAEKALLYMDGAQIDGNVVKARF 489
Query: 208 ----DQSENSKPVDSLPSRLIDQHRWPSRIGGKVSSSALNRSVDFGDARMLKTPASGTGF 263
Q + P P R D R G +V R + R K P S
Sbjct: 490 TLPPRQKASPPPKAVAPKR--DAPR-TDNAGAEVEKDGPKRPRESSPRR--KPPPSPRRR 654
Query: 264 SSL-RRLSLSEEASKPLQRASSDSVRLLSLVASGRTGSEVKSVDDCSLHDLRPHRSSTPH 322
S + RR P +RA S R L P+R
Sbjct: 655 SPVPRRAGSPRRPESPRRRADSPVRRRLD----------------------SPYRRGAGD 768
Query: 323 TDRAGLSIAGVRSQSLSAPGSRLPSPSRISVQSTSSSRGVSPSRSRPSTPPSRGGVSPSR 382
T ++ R +S S P RL SP+R+S + S G P R R PP R P R
Sbjct: 769 TPPRRRPVSPGRGRSPSPPPRRLRSPARVSPRRMRGSPG--PGRRRSPPPPPRRRSPPRR 942
Query: 383 IR 384
R
Sbjct: 943 AR 948
Score = 31.6 bits (70), Expect = 0.94
Identities = 24/72 (33%), Positives = 32/72 (44%), Gaps = 1/72 (1%)
Frame = +1
Query: 315 PHRSSTPHTDRAGLSIAGVRSQSLSAPGSRLPSPSRISVQSTSSSRG-VSPSRSRPSTPP 373
P R+ +P + R ++ S RL SP R T R VSP R R +PP
Sbjct: 664 PRRAGSPRRPESP------RRRADSPVRRRLDSPYRRGAGDTPPRRRPVSPGRGRSPSPP 825
Query: 374 SRGGVSPSRIRP 385
R SP+R+ P
Sbjct: 826 PRRLRSPARVSP 861
Score = 29.3 bits (64), Expect = 4.7
Identities = 23/46 (50%), Positives = 24/46 (52%), Gaps = 2/46 (4%)
Frame = +1
Query: 342 GSRLPSPSRISVQSTSSSRGVSPSR--SRPSTPPSRGGVSPSRIRP 385
G R P PS +S S SSSR S SR S S PSR S SR P
Sbjct: 4 GYRAP-PSSLSSSSRSSSRSRSRSRSFSSSSRSPSRSSKSRSRSPP 138
>BQ140761 weakly similar to GP|18076245|emb phosphophoryn {Rattus
norvegicus}, partial (9%)
Length = 620
Score = 43.1 bits (100), Expect = 3e-04
Identities = 49/195 (25%), Positives = 76/195 (38%), Gaps = 42/195 (21%)
Frame = +2
Query: 4 CESQQASSRRLKAVETPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGS 63
C+S+ S + PP K + T+T+++ S +R T+R ++P+ S P
Sbjct: 35 CDSRSHSHSPWPSPTRRAPP---RPKTSRPTSTSSSPSS-SRRATTRPRAPSQASPPGPP 202
Query: 64 RRCPSPN----------------------LTRTATPAA-----------------SLQLL 84
R PSP TRTATPA SL L
Sbjct: 203 SRPPSPMPWT*TRRAATAATRTAAIAVTAATRTATPATIATTAATRAQTTALRTRSLYLR 382
Query: 85 PKRSLSAERKRPSTPPSPPSRRLSTPVNDSAIDGRLS---SRKVAASRLPEGHNLWPSTM 141
P+R R+RPS P PP+ +TP ++ L SR R+P P+T
Sbjct: 383 PRRRRRRRRRRPSRSP-PPATPATTPAASPSLSPSLXTCPSRNQTTPRVP------PATR 541
Query: 142 RSLSVSFQSDIISIP 156
+L + ++ ++ P
Sbjct: 542 TALQTALRTAHLTAP 586
>AW696082
Length = 607
Score = 42.0 bits (97), Expect = 7e-04
Identities = 25/82 (30%), Positives = 42/82 (50%)
Frame = -1
Query: 530 VDIEHLKVAICQAVDVMQAMGSAIRSLFSQVEGMNDLISGVAVVATKEKSILDECEMLLA 589
VD+ + + A+ VM+ + S + L + E + IS +A V E++ ++EC L+
Sbjct: 598 VDVREVGEFLNSALKVMETIISNTQRLMPKAEETDTFISELARVVGGERAFIEECGGFLS 419
Query: 590 SVAALQVEESSLQTHLIQFKQV 611
QVEE SL+ LIQ +
Sbjct: 418 KTHKSQVEECSLRAQLIQLHSI 353
>TC84279 weakly similar to GP|1799947|dbj|BAA16433.1 BENZENE 1 2-DIOXYGENASE
ALPHA SUBUNIT (EC 1.14.12.3). {Escherichia coli},
partial (24%)
Length = 831
Score = 41.6 bits (96), Expect = 0.001
Identities = 47/158 (29%), Positives = 64/158 (39%), Gaps = 4/158 (2%)
Frame = +3
Query: 32 AATATATARRSRTREVTSRYKSPTPPSTPSGSRRCPSPNLTRTATPAASLQLLPKRSLSA 91
+++++ TAR + TR TS P+P S + + SP T TA P AS P R
Sbjct: 303 SSSSSRTARAASTRSTTSAGTGPSPSSRATAATASSSPAST-TAGPTASAATWP-RPPGT 476
Query: 92 ERKRPSTPP---SPPSRRLSTPVNDSAIDGRLSSRKVAASRLPEGHNLWPSTMRSLSVSF 148
R ST P S PS STP S R R P L ST RS S +
Sbjct: 477 RTSRASTSPRTASFPSTSTSTPTASSGSTWTPPRR----PRSPGTTTLTASTSRSASSTT 644
Query: 149 QSDIISIPVSKKER-PVTSASDRTLRPASNVAHRQGET 185
++ + R T S T ++ A+R+ T
Sbjct: 645 TLTTTTLTTRGRWRADTTGRSSPTTTTSATTANRRTRT 758
Score = 30.4 bits (67), Expect = 2.1
Identities = 30/110 (27%), Positives = 48/110 (43%)
Frame = +3
Query: 6 SQQASSRRLKAVETPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGSRR 65
++ AS+R + T P + + AATA+++ + S P PP T + SR
Sbjct: 324 ARAASTRSTTSAGTGPSP---SSRATAATASSSPASTTAGPTASAATWPRPPGTRT-SRA 491
Query: 66 CPSPNLTRTATPAASLQLLPKRSLSAERKRPSTPPSPPSRRLSTPVNDSA 115
SP RTA+ ++ P S + P P SP + L+ + SA
Sbjct: 492 STSP---RTASFPSTSTSTPTASSGSTWTPPRRPRSPGTTTLTASTSRSA 632
>BQ751767 similar to SP|O10302|GTA_N Probable global transactivator. [OpMNPV]
{Orgyia pseudotsugata multicapsid polyhedrosis virus},
partial (7%)
Length = 771
Score = 40.4 bits (93), Expect = 0.002
Identities = 55/175 (31%), Positives = 75/175 (42%), Gaps = 7/175 (4%)
Frame = +2
Query: 27 AEKNNAA--TATATARRSRTREVTSRYKS----PTPPSTPSGSRRCPSPNLTRTATPAAS 80
A+ + AA T++ TA R R SR ++ T PS PS + R + TR+A +S
Sbjct: 326 ADTSRAASPTSSTTAPAERRRWTRSRRRTWW*RRTTPSRPSSTGRNRAS--TRSAGTESS 499
Query: 81 LQLLPKRSLSAERKRPSTPPSPPSRRLSTPVNDSAIDGRLSSRKVAASRLPEGHNLWPST 140
+R+ SA R RPST P+ PSR P D A R SR + P WPS
Sbjct: 500 ST---RRTSSAARPRPSTRPAAPSR----PTRDGASPAR-PSRTGCPTLAPS----WPS- 640
Query: 141 MRSLSVSFQSDIISIPVSKKERPVTSASDRTLRPASNVAH-RQGETPSTIRKATP 194
S P R ++A+ R+ P S A R G + R +TP
Sbjct: 641 -------------SAPAPLTTRASSAATSRS--PLSRAARTRTGSREHSSRCSTP 760
Score = 34.3 bits (77), Expect = 0.14
Identities = 36/124 (29%), Positives = 50/124 (40%), Gaps = 27/124 (21%)
Frame = +2
Query: 286 SVRLLSLVASGRTGSEVKSVDDCSLHDLR-------------PHRSSTPHTDRAGLSIAG 332
S RLL A+GR S K V C DL+ P S+T +R + +
Sbjct: 224 SCRLLCSSATGRPRSTSKQVPRCPSRDLQKA*PFADTSRAASPTSSTTAPAERRRWTRSR 403
Query: 333 VRS--------QSLSAPGSRLPSPSRISVQSTSSSRGVSPSRSRPSTPPS------RGGV 378
R+ S+ G S +S+S+ R S +R RPST P+ R G
Sbjct: 404 RRTWW*RRTTPSRPSSTGRNRASTRSAGTESSSTRRTSSAARPRPSTRPAAPSRPTRDGA 583
Query: 379 SPSR 382
SP+R
Sbjct: 584 SPAR 595
>TC83962 similar to PIR|B86289|B86289 hypothetical protein AAF71991.1
[imported] - Arabidopsis thaliana, partial (11%)
Length = 679
Score = 40.4 bits (93), Expect = 0.002
Identities = 40/146 (27%), Positives = 58/146 (39%), Gaps = 1/146 (0%)
Frame = +2
Query: 27 AEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGSRRCPSPNLTRTATPAASLQLLPK 86
A K+ A + ++ + TS S +PP TPS S PSP P SL
Sbjct: 47 ANKSPA*ISNSSTPSPSSSTSTSTSSSSSPPPTPSTSSTTPSP------PPPTSLSSPTP 208
Query: 87 RSLSAERKRPSTPPSPPSRRLSTPVNDSAIDGRLSSRKVAASRLPEGHNLWPSTMRSL-S 145
S P+TP + + STP+N + L + S LP + +T +L S
Sbjct: 209 TSNHLSSA*PTTPTNTLTAVPSTPLNSPCSNLHLPQQTSLPSPLPSSSPSYQTTPPALAS 388
Query: 146 VSFQSDIISIPVSKKERPVTSASDRT 171
VS S +P+ SA +T
Sbjct: 389 VSLSSSATPLPLPAHSLVNISAYSQT 466
Score = 30.4 bits (67), Expect = 2.1
Identities = 22/51 (43%), Positives = 26/51 (50%), Gaps = 1/51 (1%)
Frame = +2
Query: 343 SRLPSPSRISVQSTSSSRGVSPSRSRPSTPPS-RGGVSPSRIRPTSSSIQS 392
S PSPS + STSSS P+ S ST PS S S PTS+ + S
Sbjct: 77 SSTPSPSSSTSTSTSSSSSPPPTPSTSSTTPSPPPPTSLSSPTPTSNHLSS 229
>BQ751262 similar to GP|6715100|gb|A versicolorin B synthase {Aspergillus
parasiticus}, partial (11%)
Length = 710
Score = 40.0 bits (92), Expect = 0.003
Identities = 37/119 (31%), Positives = 49/119 (41%), Gaps = 1/119 (0%)
Frame = +3
Query: 19 TPRPPLVLAEKNNAATATATARRSRTREVT-SRYKSPTPPSTPSGSRRCPSPNLTRTATP 77
+PRP AE + A + T R + + +R +S PS SR PSP+
Sbjct: 396 SPRPRTSTAESSWARSTAPTPSPQRRKSASRARRRSSPKPSAARTSRSTPSPS------- 554
Query: 78 AASLQLLPKRSLSAERKRPSTPPSPPSRRLSTPVNDSAIDGRLSSRKVAASRLPEGHNL 136
P+RS S KRP SPP+ ++P S R SS A S P H L
Sbjct: 555 -------PRRSSSTATKRPPASSSPPTP--ASPPTPSR-RARRSSSPPAPSEPPAPHGL 701
Score = 37.7 bits (86), Expect = 0.013
Identities = 40/162 (24%), Positives = 64/162 (38%), Gaps = 2/162 (1%)
Frame = +3
Query: 230 PSRIGGKVSSSALNRSVDFGDARMLKTPASGTGFSSLRRLSLSEEASKPLQRASSDSVRL 289
P+R G +S++ L L +P S SSL R+ P + ++
Sbjct: 159 PTRSGRTLSATTLTHGT-------LSSPTSRRASSSLPRVLRDSPTPAP------STTQM 299
Query: 290 LSLVASGRTGSEVKSVDDCSLHDLRP--HRSSTPHTDRAGLSIAGVRSQSLSAPGSRLPS 347
S +G S + ++ S H P RS++P + + RS + + R S
Sbjct: 300 PSPPPAGLLRSRMPTMPSRSAHISSPLSTRSASPRPRTSTAESSWARSTAPTPSPQRRKS 479
Query: 348 PSRISVQSTSSSRGVSPSRSRPSTPPSRGGVSPSRIRPTSSS 389
SR +S+ SRS PS P R + ++ P SSS
Sbjct: 480 ASRARRRSSPKPSAARTSRSTPSPSPRRSSSTATKRPPASSS 605
Score = 30.8 bits (68), Expect = 1.6
Identities = 22/100 (22%), Positives = 41/100 (41%)
Frame = +3
Query: 7 QQASSRRLKAVETPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGSRRC 66
++ S+ R + +P+P ++ + + + + T+ + PTP S P+ SRR
Sbjct: 468 RRKSASRARRRSSPKPSAARTSRSTPSPSPRRSSSTATKRPPASSSPPTPASPPTPSRRA 647
Query: 67 PSPNLTRTATPAASLQLLPKRSLSAERKRPSTPPSPPSRR 106
R+++P A PS PP+P R
Sbjct: 648 -----RRSSSPPA----------------PSEPPAPHGLR 704
>TC81100 similar to PIR|H84824|H84824 En/Spm-like transposon protein
[imported] - Arabidopsis thaliana, partial (20%)
Length = 1266
Score = 40.0 bits (92), Expect = 0.003
Identities = 51/195 (26%), Positives = 89/195 (45%), Gaps = 16/195 (8%)
Frame = +3
Query: 26 LAEKNNAATATATARRSRTREVTSRYKSPTPP----STPSGSRRCPSPNLT-RTATPAAS 80
+ K++A+ ++ + + ++ ++S + P+P STPSG PS + R +TP +
Sbjct: 654 VVSKHHASMSSFGSSTNGSKRISSS-RGPSPATSRSSTPSGRPTLPSTTKSSRPSTPTSR 830
Query: 81 LQLLPKRSLSAERKRPSTPPSPPSRRLSTPVNDSAIDGRLSSRKVAASRLPEGHNLWPST 140
L +S +A R STP P S R STP + ++ +S++ S P + PS
Sbjct: 831 ATLTSTKS-TAPPVRSSTPTRPTS-RASTPTSRPSLTAPKTSQR---SATPNIRSSTPS- 992
Query: 141 MRSLSVSFQSDIISIPVSKKERPVTS---ASDRTLRPA--------SNVAHRQGETPSTI 189
R+ VS S + K RPV + R + P+ S + + PS +
Sbjct: 993 -RTFGVSAPPTRPS--STSKARPVVAKNPVQSRGISPSVKSRPWEPSQMPGFTHDAPSNL 1163
Query: 190 RKATPERKRSPLKGK 204
+K+ PER S + +
Sbjct: 1164KKSLPERPASVTRNR 1208
Score = 34.7 bits (78), Expect = 0.11
Identities = 30/79 (37%), Positives = 41/79 (50%), Gaps = 6/79 (7%)
Frame = +3
Query: 317 RSSTPHTDRAGLSIAG-----VRSQSLSAPGSRLPSP-SRISVQSTSSSRGVSPSRSRPS 370
R STP T RA L+ VRS + + P SR +P SR S+ + +S+ + R S
Sbjct: 807 RPSTP-TSRATLTSTKSTAPPVRSSTPTRPTSRASTPTSRPSLTAPKTSQRSATPNIRSS 983
Query: 371 TPPSRGGVSPSRIRPTSSS 389
TP GVS RP+S+S
Sbjct: 984 TPSRTFGVSAPPTRPSSTS 1040
Score = 33.1 bits (74), Expect = 0.32
Identities = 56/216 (25%), Positives = 85/216 (38%), Gaps = 6/216 (2%)
Frame = +3
Query: 55 TPPSTP---SGSRRCPSPNLTRTATPAASLQLLPKRSLSAERKRPSTPPSPPSRRLSTPV 111
TPP TP + + P + T T ++ L R + + + S S
Sbjct: 507 TPPDTPLFPTLDKESQMPLKSETETHSSRPTALKLRVDNIQIEPSSRSNVVSKHHASMSS 686
Query: 112 NDSAIDGRLSSRKVAASRLPEGHNLWPSTMRSLSVSFQSDIISIPVSKKERPVTSASDRT 171
S+ +G S+++++SR P P+T RS + S + + S +K RP T S T
Sbjct: 687 FGSSTNG---SKRISSSRGPS-----PATSRSSTPSGRPTLPS--TTKSSRPSTPTSRAT 836
Query: 172 LRPASNVAHR-QGETPS--TIRKATPERKRSPLKGKNASDQSENSKPVDSLPSRLIDQHR 228
L + A + TP+ T R +TP R L S +S S PSR
Sbjct: 837 LTSTKSTAPPVRSSTPTRPTSRASTP-TSRPSLTAPKTSQRSATPNIRSSTPSRTFGVSA 1013
Query: 229 WPSRIGGKVSSSALNRSVDFGDARMLKTPASGTGFS 264
P+R SS++ R V + K P G S
Sbjct: 1014PPTR----PSSTSKARPV------VAKNPVQSRGIS 1091
Score = 30.8 bits (68), Expect = 1.6
Identities = 42/145 (28%), Positives = 58/145 (39%), Gaps = 4/145 (2%)
Frame = +3
Query: 242 LNRSVDFGDARMLKTPASGTGFSSLRRLSLSEEASKPLQRASSDSVRLLSLVASGRTGSE 301
LN D D L TP F +L +E+ PL+ + S R +
Sbjct: 465 LNSENDKSDYEWLLTPPDTPLFPTL-----DKESQMPLKSETETH--------SSRPTAL 605
Query: 302 VKSVDDCSLHDLRPHRSSTPHTDRAGLSIAGVRSQ-SLSAPGSRLPSPSRISVQSTSSSR 360
VD+ + RS+ A +S G + S SR PSP+ S ST S R
Sbjct: 606 KLRVDNIQIEP--SSRSNVVSKHHASMSSFGSSTNGSKRISSSRGPSPAT-SRSSTPSGR 776
Query: 361 GVSPSR---SRPSTPPSRGGVSPSR 382
PS SRPSTP SR ++ ++
Sbjct: 777 PTLPSTTKSSRPSTPTSRATLTSTK 851
Score = 30.8 bits (68), Expect = 1.6
Identities = 44/147 (29%), Positives = 64/147 (42%), Gaps = 17/147 (11%)
Frame = +3
Query: 260 GTGFSSLRRLSLSEEASKPLQRASSDSVR--LLSLVASGRTGSEVKSVDDCSLHDLRPH- 316
G+ + +R+S S S R+S+ S R L S S R + S P
Sbjct: 690 GSSTNGSKRISSSRGPSPATSRSSTPSGRPTLPSTTKSSRPSTPTSRATLTSTKSTAPPV 869
Query: 317 RSSTPHTDRAGLSIAGVRSQSLSAPGSRLPSPSRISVQSTSSSR--GVSPSRSRPST--- 371
RSSTP + S R SL+AP + S + +++S++ SR GVS +RPS+
Sbjct: 870 RSSTPTRPTSRASTPTSRP-SLTAPKTSQRSATP-NIRSSTPSRTFGVSAPPTRPSSTSK 1043
Query: 372 --------PPSRGGVSPS-RIRPTSSS 389
P G+SPS + RP S
Sbjct: 1044ARPVVAKNPVQSRGISPSVKSRPWEPS 1124
>BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xylostella
granulovirus}, partial (42%)
Length = 909
Score = 39.7 bits (91), Expect = 0.003
Identities = 28/101 (27%), Positives = 40/101 (38%), Gaps = 2/101 (1%)
Frame = +3
Query: 19 TPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPP--STPSGSRRCPSPNLTRTAT 76
TP P K + + T R + + + R K PP +TP + P P T T
Sbjct: 18 TPSPQATPQRKGHPSQKPPTGRSHKKTKKSPRKKEKPPPRPATPPPAPAPPGPPNQTTNT 197
Query: 77 PAASLQLLPKRSLSAERKRPSTPPSPPSRRLSTPVNDSAID 117
Q P + RP TPP+PP +TP ++ D
Sbjct: 198 TNPKKQHTP--TTPTHPPRPHTPPTPPPTTQTTPPREAPTD 314
Score = 29.3 bits (64), Expect = 4.7
Identities = 18/57 (31%), Positives = 26/57 (45%)
Frame = +1
Query: 16 AVETPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGSRRCPSPNLT 72
A RPP EKN AA A ++ +T + + + PP TP ++ P P T
Sbjct: 736 ATSRKRPPRGAREKNPAAHAREREKK-KTPKKNPQNHNNNPPKTPPPQKKHPPPPKT 903
>TC80891 similar to GP|21622328|emb|CAD36969. probable isoamyl alcohol
oxidase {Neurospora crassa}, partial (12%)
Length = 766
Score = 39.3 bits (90), Expect = 0.004
Identities = 29/98 (29%), Positives = 48/98 (48%), Gaps = 1/98 (1%)
Frame = +1
Query: 19 TPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGSRRCPSPNLTRTATPA 78
+P P L++ + ++ + +R R++ S + PS P SR PS N ++
Sbjct: 154 SPSPHLLIPSSLSLSSPFSFSRWPRSQRWC--LSSTSSPSPPRRSRPLPSENPGAPSSEE 327
Query: 79 ASLQLLPKRSLSAERKRPSTPPS-PPSRRLSTPVNDSA 115
L+ + + A RPS PPS PPS +S+P + SA
Sbjct: 328 TRLRAVASPATPAGPPRPSGPPSTPPSAAVSSPPSPSA 441
>TC84022 homologue to GP|15822513|gb|AAG23338.1 thiamine biosynthesis
protein NMT-1 {Neurospora crassa}, partial (46%)
Length = 613
Score = 38.9 bits (89), Expect = 0.006
Identities = 29/95 (30%), Positives = 41/95 (42%), Gaps = 14/95 (14%)
Frame = +2
Query: 42 SRTREVTSRYKSPTPPS-TPSGSRRC-------------PSPNLTRTATPAASLQLLPKR 87
SRTR S + SPT P+ +P S P T + P+A+ P R
Sbjct: 227 SRTRASRSLFSSPTTPAMSPKSSEPARLTSASRP*FTLSPPRRATSPSCPSAASSTSPSR 406
Query: 88 SLSAERKRPSTPPSPPSRRLSTPVNDSAIDGRLSS 122
+ S R R S P S PSR ++ + S+ R +S
Sbjct: 407 ASSTSRTRASRPTSAPSRASASATSASSARSRSTS 511
>TC93234 weakly similar to GP|21751020|dbj|BAC03887. unnamed protein product
{Homo sapiens}, partial (11%)
Length = 591
Score = 38.1 bits (87), Expect = 0.010
Identities = 53/179 (29%), Positives = 74/179 (40%), Gaps = 6/179 (3%)
Frame = +2
Query: 6 SQQASSRRLKAVETPRPPLVLAEKNNAATATATARRSRT-REVTSRYKSPT----PPSTP 60
SQQ SR + E P A + ++++ ++RSRT R +SR + + PP
Sbjct: 110 SQQKWSRTS*SSERATP----ASASLTSSSSIPSQRSRTSRSPSSRPRHTSTGTAPPFAV 277
Query: 61 SGSRRCPS-PNLTRTATPAASLQLLPKRSLSAERKRPSTPPSPPSRRLSTPVNDSAIDGR 119
S P+ P+ TR++ + S P S SA R PSTPP P R S P S+
Sbjct: 278 SSPASSPTTPSSTRSSPGSTSTPRTPSTSFSA-RPPPSTPPPTPFR--SRPTKGSSPSST 448
Query: 120 LSSRKVAASRLPEGHNLWPSTMRSLSVSFQSDIISIPVSKKERPVTSASDRTLRPASNV 178
SS A P + PS S P + P T+ S R+ P S V
Sbjct: 449 PSSSSPPA---PTSPAISPSRP------------SAPTRRPSPPGTACSPRSRPPNSIV 580
Score = 33.9 bits (76), Expect = 0.19
Identities = 32/101 (31%), Positives = 43/101 (41%), Gaps = 5/101 (4%)
Frame = +2
Query: 298 TGSEVKSVDDCSLHDLRPHRSSTPHTDRAGLSIAGVRSQSLSAPGSR--LPSPSRISVQS 355
T + S+ R S +P + S ++S+P S PS +R S S
Sbjct: 155 TPASASLTSSSSIPSQRSRTSRSPSSRPRHTSTGTAPPFAVSSPASSPTTPSSTRSSPGS 334
Query: 356 TSSSRGVSPS---RSRPSTPPSRGGVSPSRIRPTSSSIQSN 393
TS+ R S S R PSTPP +P R RPT S S+
Sbjct: 335 TSTPRTPSTSFSARPPPSTPPP----TPFRSRPTKGSSPSS 445
>BQ137210 similar to SP|P09789|GRP1_ Glycine-rich cell wall structural
protein 1 precursor. [Petunia] {Petunia hybrida},
partial (13%)
Length = 990
Score = 38.1 bits (87), Expect = 0.010
Identities = 34/116 (29%), Positives = 47/116 (40%), Gaps = 10/116 (8%)
Frame = -2
Query: 5 ESQQASSRRLKAVETPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGSR 64
+ + + R +A E P L A + + T RR TR + ++P SR
Sbjct: 524 QRDHSETTRARAPERRPPHLARAHR----PTSRTTRRHATRACRTHTRAPP-------SR 378
Query: 65 RCPSPNLTRTATPAASLQLLPKRSLSAERK----------RPSTPPSPPSRRLSTP 110
R P P T T SL P+R + K RP PPSPP+R +TP
Sbjct: 377 RRPGPAFT-PQTRECSLHSDPQRPQKKQSKKTNEPTQLFPRPLVPPSPPTRLAATP 213
>BQ752298 similar to GP|11595639|em conserved hypothetical protein
{Neurospora crassa}, partial (2%)
Length = 834
Score = 37.7 bits (86), Expect = 0.013
Identities = 34/122 (27%), Positives = 45/122 (36%), Gaps = 15/122 (12%)
Frame = -1
Query: 27 AEKNNAATATATARRSRTREVTSRYKSP------------TPPSTPSGSRRCPSPNLTRT 74
AE A TAT R+ R T P PP PSG+ P++
Sbjct: 633 AEATQAKTATPPPPRAAVRSTTGTPPPPHA*MTPPMGRASAPPGGPSGTELNKPPSMESL 454
Query: 75 ATPAASLQLLPKRSLSAERKRPSTPPSPPSRRLSTPVNDSAIDGRLSS---RKVAASRLP 131
P RS+S PPSR ++ N S+ID LS+ RK A +
Sbjct: 453 GAPPGPRPPSMMRSVSNTSAAGGPLSGPPSRPATSMSNASSIDDLLSAAGPRKPGAKKAK 274
Query: 132 EG 133
+G
Sbjct: 273 KG 268
>BF638475 similar to GP|9502414|gb| peroxisomal targeting signal type 2
receptor Pex7p {Arabidopsis thaliana}, partial (45%)
Length = 511
Score = 37.7 bits (86), Expect = 0.013
Identities = 29/87 (33%), Positives = 39/87 (44%)
Frame = +1
Query: 303 KSVDDCSLHDLRPHRSSTPHTDRAGLSIAGVRSQSLSAPGSRLPSPSRISVQSTSSSRGV 362
KS C L+PH + TP S SA +++ SPS S SS+ V
Sbjct: 40 KSFKRCQF--LKPHSTVTP---------------SNSALSTKIASPSPPLKTSASSATAV 168
Query: 363 SPSRSRPSTPPSRGGVSPSRIRPTSSS 389
S S + P PPS+ SP PT+S+
Sbjct: 169 STSSNSPPIPPSQSPKSPLTTPPTAST 249
>TC91032 weakly similar to GP|20196995|gb|AAB91977.2 expressed protein
{Arabidopsis thaliana}, partial (37%)
Length = 1366
Score = 37.4 bits (85), Expect = 0.017
Identities = 31/91 (34%), Positives = 42/91 (46%), Gaps = 14/91 (15%)
Frame = -2
Query: 28 EKNNAATATATARRSRTREVTSRYKSP----TPPSTPSGSRRCPSPNLTRT-----ATPA 78
+ N++ +T T S T T+ P TPPSTPS + PS + + T T
Sbjct: 603 KSNSSLQSTETIHFSTTHTPTALVHLPPQFSTPPSTPSTLPQQPSTSHSSTLPSHSPTSH 424
Query: 79 ASLQLLPK-----RSLSAERKRPSTPPSPPS 104
++LQ LP S S PS+PP PPS
Sbjct: 423 STLQSLPPTLTTLSSPSTPHFSPSSPPQPPS 331
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.312 0.125 0.345
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,714,724
Number of Sequences: 36976
Number of extensions: 248673
Number of successful extensions: 2730
Number of sequences better than 10.0: 229
Number of HSP's better than 10.0 without gapping: 2156
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2572
length of query: 616
length of database: 9,014,727
effective HSP length: 102
effective length of query: 514
effective length of database: 5,243,175
effective search space: 2694991950
effective search space used: 2694991950
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0037a.4


