

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
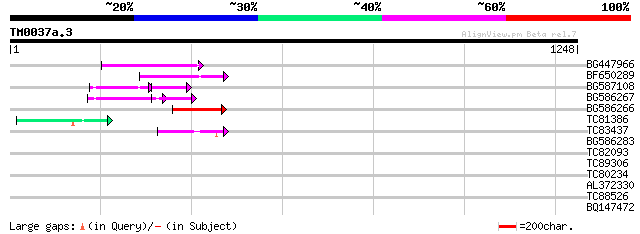
Query= TM0037a.3
(1248 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [impo... 150 4e-36
BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgari... 134 2e-31
BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - ... 79 3e-27
BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At... 103 3e-22
BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-li... 103 4e-22
TC81386 similar to GP|10140689|gb|AAG13524.1 putative non-LTR re... 76 7e-14
TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imp... 72 1e-12
BG586283 similar to GP|7267666|em RNA-directed DNA polymerase-li... 35 0.24
TC82093 34 0.31
TC89306 similar to SP|P56820|IF37_ARATH Eukaryotic translation i... 32 1.5
TC80234 similar to GP|12325097|gb|AAG52506.1 unknown protein; 31... 32 2.0
AL372330 similar to GP|21322752|dbj cold shock protein-1 {Tritic... 30 4.5
TC88526 similar to PIR|T49291|T49291 hypothetical protein T16L24... 29 10.0
BQ147472 similar to GP|21555880|gb| unknown {Arabidopsis thalian... 29 10.0
>BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [imported] -
Arabidopsis thaliana, partial (7%)
Length = 687
Score = 150 bits (378), Expect = 4e-36
Identities = 84/225 (37%), Positives = 118/225 (52%), Gaps = 1/225 (0%)
Frame = +2
Query: 202 WLNHGDKNTRFFHTSTMVRRKRNKIEALTDEHGVSVTDPITLRSMAIDFFQHLYTSPGTD 261
WL GDKNT+FFH+ RRK N+I+ L DE G + + I +F +L+TS
Sbjct: 5 WLKDGDKNTKFFHSKASQRRKVNEIKKLKDETGNWCKGEENVERLLITYFNNLFTSSNPT 184
Query: 262 VLYHVRNAFP-TLPRDTLLEISNPLEADEIKAAVFSMRALKAPGPDGLNTLFFQSQWDTI 320
+ L + ++ +E+ A+ M +KAPGPDGL LFFQ W +
Sbjct: 185 AIEETCEVVKGKLSHEHIVWCEKEFTEEEVLEAINQMHPVKAPGPDGLPALFFQKYWHIV 364
Query: 321 GSSVVQFIQDCIQRPEKIREVNDTLIVLIPKIDRPNLLSQYRLIGICNVIYKTLTKALAN 380
G V Q + + + E+N T IVLIPK PN YR I +CNV+ K +TK +AN
Sbjct: 365 GKEVQQMVLQVLNNSMETEELNKTFIVLIPKGKNPNTPKDYRPISLCNVVMKIITKVIAN 544
Query: 381 RLKGVLGDLISPNQCSFIPGRQSSDNIIIAQEVFHSMRYLKRKKG 425
R+K L D+I Q +F+ GR +DN +IA V S+ R+KG
Sbjct: 545 RVKQTLPDVIDVEQSAFVQGRLITDNALIAWSV--SIG*RXRRKG 673
>BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgaris}, partial
(69%)
Length = 616
Score = 134 bits (337), Expect = 2e-31
Identities = 74/194 (38%), Positives = 111/194 (57%)
Frame = +3
Query: 287 ADEIKAAVFSMRALKAPGPDGLNTLFFQSQWDTIGSSVVQFIQDCIQRPEKIREVNDTLI 346
A E+K A+FSM + KAPG DG N FF+ W+ IG SV+ I D + + +N T +
Sbjct: 30 AVEVKNALFSMDSSKAPGIDGYNVHFFKCSWNIIGDSVIDAILDFFKTGFMPKIINCTYV 209
Query: 347 VLIPKIDRPNLLSQYRLIGICNVIYKTLTKALANRLKGVLGDLISPNQCSFIPGRQSSDN 406
L+PK + +R I C+VIYK ++K L +R++GVL ++S NQ +F+ GR DN
Sbjct: 210 TLLPKEVNVTSVKNFRPIACCSVIYKIISKILTSRMQGVLNSVVSENQSAFVKGRVIFDN 389
Query: 407 IIIAQEVFHSMRYLKRKKGWIAIKIDLEKAYDRLEWSFLKDTLLEVGLDRNFCDLILDCV 466
II++ E+ S K +KIDL KAYD EW F+K +LE+G F + ++ +
Sbjct: 390 IILSHELVKSYS-RKGISPRCMVKIDLXKAYDSXEWPFIKHLMLELGFPYKFVNWVMAXL 566
Query: 467 SLSSFQVLFNGSKT 480
+ +S+ NG T
Sbjct: 567 TTASYTFNXNGDLT 608
>BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - Arabidopsis
thaliana, partial (17%)
Length = 677
Score = 79.0 bits (193), Expect(2) = 3e-27
Identities = 49/145 (33%), Positives = 77/145 (52%), Gaps = 4/145 (2%)
Frame = +2
Query: 175 LQKRLWKEYQTTLIQEELLWRQKSRVAWLNHGDKNTRFFHTSTMVRRKRNKIEALTDEHG 234
L+++L + Y+ EE W+QKSR W GD N +F+H T R RN+I L D G
Sbjct: 8 LKEKLQEAYK----DEEDYWQQKSRNMWHISGDLNKKFYHALTKQRHARNRIVGLYDYDG 175
Query: 235 VSVTDPITLRSMAIDFFQHLY---TSPGTD-VLYHVRNAFPTLPRDTLLEISNPLEADEI 290
+T+ + +A+D+F+ L+ T G D L + ++ LL ++ +E+
Sbjct: 176 NWITEEQGVEKVAVDYFEDLFQRTTPTGFDGFLDEITSSITPQMNQRLLRLAT---EEEV 346
Query: 291 KAAVFSMRALKAPGPDGLNTLFFQS 315
+ A+F M KAPGPDG+ TL F +
Sbjct: 347 RLALFIMHPEKAPGPDGMTTLLFST 421
Score = 62.8 bits (151), Expect(2) = 3e-27
Identities = 33/89 (37%), Positives = 48/89 (53%)
Frame = +3
Query: 312 FFQSQWDTIGSSVVQFIQDCIQRPEKIREVNDTLIVLIPKIDRPNLLSQYRLIGICNVIY 371
FFQ W I +++ + + + +N T I LIPK RP +++ R I +CNV Y
Sbjct: 411 FFQHSWHIIKMDLLKMVNSFLASGKLDTRLNTTNICLIPKKKRPTRMTELRPISLCNVGY 590
Query: 372 KTLTKALANRLKGVLGDLISPNQCSFIPG 400
K ++K L RLK L LIS Q +F+ G
Sbjct: 591 KIISKVLCQRLKVCLPSLISETQSAFVHG 677
>BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (16%)
Length = 794
Score = 103 bits (258), Expect = 3e-22
Identities = 58/176 (32%), Positives = 90/176 (50%), Gaps = 1/176 (0%)
Frame = +3
Query: 172 LEKLQKRLWKEYQTTLIQEELLWRQKSRVAWLNHGDKNTRFFHTSTMVRRKRNKIEALTD 231
L L+ L +EY EE+ W QKSR+ WL GD+NT+FFH T RR +N+I +L D
Sbjct: 81 LSLLRSELNEEYHN----EEIFWMQKSRLNWLRSGDRNTKFFHAVTKNRRAQNRILSLID 248
Query: 232 EHGVSVTDPITLRSMAIDFFQHLYTSPGTDVLYHVRNAFPTL-PRDTLLEISNPLEADEI 290
+ L +A F+ LY+S + N+ P + + ++ + +E+
Sbjct: 249 DDDKEWFVEEDLGRLADSHFKLLYSSEDVGITLEDWNSIPAIVTEEQNAQLMAQISREEV 428
Query: 291 KAAVFSMRALKAPGPDGLNTLFFQSQWDTIGSSVVQFIQDCIQRPEKIREVNDTLI 346
+ AVF + K PGPDG+N FFQ WDT+G + Q+ ++ + +N T I
Sbjct: 429 REAVFDINPHKCPGPDGMNVFFFQQFWDTMGDDLTSMAQEFLRTGKLEEGINKTNI 596
Score = 50.8 bits (120), Expect = 3e-06
Identities = 33/98 (33%), Positives = 52/98 (52%)
Frame = +1
Query: 313 FQSQWDTIGSSVVQFIQDCIQRPEKIREVNDTLIVLIPKIDRPNLLSQYRLIGICNVIYK 372
F +QW+ I + ++ ++ RE L+PK L ++R I +CNV YK
Sbjct: 505 FGTQWEMISHL---WRKNSSEQGNLKRESTKQTSGLVPKKLEAKRLVEFRPISLCNVAYK 675
Query: 373 TLTKALANRLKGVLGDLISPNQCSFIPGRQSSDNIIIA 410
++K L+ RLK VL +I+ Q +F + SDNI+IA
Sbjct: 676 IVSKVLSKRLKSVLPWIITETQAAFGRRQLISDNILIA 789
>BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 789
Score = 103 bits (257), Expect = 4e-22
Identities = 49/121 (40%), Positives = 77/121 (63%), Gaps = 1/121 (0%)
Frame = -3
Query: 358 LSQYRLIGICNVIYKTLTKALANRLKGVLGDLISPNQCSFIPGRQSSDNIIIAQEVFHSM 417
+S+YR I CN YK + K L+ R++ +L +ISP+Q +F+PGR SDN++I ++ H +
Sbjct: 778 VSEYRTIAPCNTQYKIIAKILSKRMQPLLRSIISPSQSAFVPGRAISDNVLITHKILHYL 599
Query: 418 RYLKRKKG-WIAIKIDLEKAYDRLEWSFLKDTLLEVGLDRNFCDLILDCVSLSSFQVLFN 476
R KK +A+K D+ KAYDR+ W+FL++ L +G + I++CVS S+ L N
Sbjct: 598 RQSGAKKHVSMAVKTDMTKAYDRIAWNFLREVLTRLGFHGIWISWIMECVSTVSYSFLIN 419
Query: 477 G 477
G
Sbjct: 418 G 416
>TC81386 similar to GP|10140689|gb|AAG13524.1 putative non-LTR retroelement
reverse transcriptase {Oryza sativa (japonica
cultivar-group)}, partial (1%)
Length = 798
Score = 76.3 bits (186), Expect = 7e-14
Identities = 67/266 (25%), Positives = 105/266 (39%), Gaps = 54/266 (20%)
Frame = -1
Query: 15 RNFLWQELRRIAANTGEPWVVMGDFNTYLNAPDKWGGDPPNLLAMGKFRDCLDDCYLSDL 74
R LW + I W +GDFNT L + + G P L M F+ D L L
Sbjct: 795 RRQLWSAISNIQTQHKLSWCCIGDFNTILGSHEHQGSHTPARLPMLDFQQWSDVNNLFHL 616
Query: 75 GFKGPPFTW-EGR----GVKERLDWALGNDQWLRSFPEASIFHLPQLKSDHKPLLLQLSP 129
+G FTW GR ++RLD ++ N + + S L +L+SDH P+L +L
Sbjct: 615 PTRGSAFTWTNGRRGRNNTRKRLDRSIVNQLMIDNCESLSACTLTKLRSDHFPILFELQT 436
Query: 130 IDIDHSQ----------RP------------------------------------NRQVF 143
+I S P N+ F
Sbjct: 435 QNIQFSSSFKFMKMWSAHPDCINIIKQCWANQVVGCPMFVLNQKLKNLKEVLKVWNKNTF 256
Query: 144 GEIGRRKRHLMRRLEGINSRLRMQHIPYLEKL---QKRLWKEYQTTLIQEELLWRQKSRV 200
G + + + + L+ I ++++ I Y + L +K ++ L EE+ W +KS+V
Sbjct: 255 GNVHSQVDNAYKELDDI--QVKIDSIGYSDVLMDQEKAAQLNLESALNIEEVFWHEKSKV 82
Query: 201 AWLNHGDKNTRFFHTSTMVRRKRNKI 226
W GD+NT FFH ++R + I
Sbjct: 81 NWHCEGDRNTAFFHRVAKIKRTSSLI 4
>TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imported] -
Arabidopsis thaliana, partial (6%)
Length = 951
Score = 72.0 bits (175), Expect = 1e-12
Identities = 51/166 (30%), Positives = 83/166 (49%), Gaps = 11/166 (6%)
Frame = +2
Query: 326 QFIQDCIQRPEKIREVNDTLIVLIPKIDRPNLLSQYRLIGICNVIYKTLTKALANRLKGV 385
+F+ + + + + +N T I LIPK+D P L+ +R I + +YK L K LANRL+ V
Sbjct: 5 RFVSEFHRNRKLFKGINSTFIALIPKVDNPQRLNDFRPISLVGSLYKILGKLLANRLRVV 184
Query: 386 LGDLISPNQCSFIPGRQSSDNIIIAQEVFHSMRYLKRKKGWIAIKIDLEKAYDRLEWSFL 445
+G +IS Q +F+ RQ + M +L + + W+ ++ + K + L W
Sbjct: 185 IGSVISDAQSAFVKNRQILE-----------MVFL*QMRLWMRLR-N*RKIFCCLRWILK 328
Query: 446 KDTLLEVGL-------DRNFCDL----ILDCVSLSSFQVLFNGSKT 480
+ L +GL +F L I +CVS ++ VL NGS T
Sbjct: 329 RLITLSIGLIWILF*VGMSFLVLWRKWIKECVSTATTSVLVNGSPT 466
>BG586283 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (7%)
Length = 774
Score = 34.7 bits (78), Expect = 0.24
Identities = 17/44 (38%), Positives = 21/44 (47%)
Frame = +3
Query: 8 GSPNLSIRNFLWQELRRIAANTGEPWVVMGDFNTYLNAPDKWGG 51
G P R W EL I A W++ GDFN L+ +K GG
Sbjct: 519 GVPQPEHRAKFWDELSTIGAQRDGAWLLTGDFNDLLDNSEKVGG 650
>TC82093
Length = 569
Score = 34.3 bits (77), Expect = 0.31
Identities = 14/28 (50%), Positives = 19/28 (67%)
Frame = -1
Query: 187 LIQEELLWRQKSRVAWLNHGDKNTRFFH 214
L Q E+LW KS+ W+ +G +NT FFH
Sbjct: 473 LSQNEMLWY*KSQ*NWVKYGSRNTWFFH 390
>TC89306 similar to SP|P56820|IF37_ARATH Eukaryotic translation initiation
factor 3 subunit 7 (eIF-3 zeta) (eIF3d) (p66). [Mouse-ear
cress], partial (45%)
Length = 1037
Score = 32.0 bits (71), Expect = 1.5
Identities = 35/122 (28%), Positives = 58/122 (46%), Gaps = 7/122 (5%)
Frame = -1
Query: 268 NAFPTLPRDTLLEISN--PLEADEI-KAAVFSMRALKAPGPDGLNTLFFQSQWDTIGSSV 324
N FP + + L SN P+ + + + +R+L A G G+N +F QW + S+
Sbjct: 1031 NLFPFISKRIWLIKSNLFPIPNQNLXREILIDIRSLHAKGMSGINIIFSLRQW--LLRSL 858
Query: 325 VQFIQDCIQRPEKIRE--VNDTLIVLIPKIDRPNLLSQYR--LIGICNVIYKTLTKALAN 380
+ Q ++ I+E V +TL IP+++RP Q R IG N + + K N
Sbjct: 857 MDRKQIQLRPITLIKEKLVPNTLNNNIPRVNRPRSTHQRR*NSIGSKNRSFILICKPTNN 678
Query: 381 RL 382
R+
Sbjct: 677 RV 672
>TC80234 similar to GP|12325097|gb|AAG52506.1 unknown protein; 31958-32770
{Arabidopsis thaliana}, partial (65%)
Length = 1374
Score = 31.6 bits (70), Expect = 2.0
Identities = 14/40 (35%), Positives = 20/40 (50%)
Frame = +2
Query: 442 WSFLKDTLLEVGLDRNFCDLILDCVSLSSFQVLFNGSKTE 481
W+F L +G+ R+ + IL C S +V N SK E
Sbjct: 296 WNFASHALTSIGMKRSSTEPILSCAECSDDEVCSNASKDE 415
>AL372330 similar to GP|21322752|dbj cold shock protein-1 {Triticum
aestivum}, partial (33%)
Length = 435
Score = 30.4 bits (67), Expect = 4.5
Identities = 18/60 (30%), Positives = 28/60 (46%), Gaps = 5/60 (8%)
Frame = +1
Query: 148 RRKRHLMRRLEGINSRLRMQHIPYLEKLQKRLWKEYQ-----TTLIQEELLWRQKSRVAW 202
R +R RRL I RLR ++Q+R+W+ + + L WRQ+ R+ W
Sbjct: 100 RLRRRFERRLRRIQRRLR--------RIQRRIWRRRVWWRSIRRIWRRRLRWRQRRRIWW 255
>TC88526 similar to PIR|T49291|T49291 hypothetical protein T16L24.50 -
Arabidopsis thaliana, partial (96%)
Length = 1064
Score = 29.3 bits (64), Expect = 10.0
Identities = 10/20 (50%), Positives = 16/20 (80%)
Frame = +1
Query: 835 GKLISSSTFYLLLPWRRLCL 854
GK+IS ++Y+L+PW LC+
Sbjct: 703 GKIISGYSYYVLMPWFCLCM 762
>BQ147472 similar to GP|21555880|gb| unknown {Arabidopsis thaliana},
partial (29%)
Length = 598
Score = 29.3 bits (64), Expect = 10.0
Identities = 11/29 (37%), Positives = 18/29 (61%), Gaps = 3/29 (10%)
Frame = +1
Query: 33 WVVMGDFNTYLN---APDKWGGDPPNLLA 58
W + GDF++Y++ P WGG+P +A
Sbjct: 55 WFIEGDFDSYISQIRKPHVWGGEPELFIA 141
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.343 0.152 0.539
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 44,265,617
Number of Sequences: 36976
Number of extensions: 701212
Number of successful extensions: 4258
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 2306
Number of HSP's successfully gapped in prelim test: 175
Number of HSP's that attempted gapping in prelim test: 1865
Number of HSP's gapped (non-prelim): 2626
length of query: 1248
length of database: 9,014,727
effective HSP length: 107
effective length of query: 1141
effective length of database: 5,058,295
effective search space: 5771514595
effective search space used: 5771514595
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0037a.3


