

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
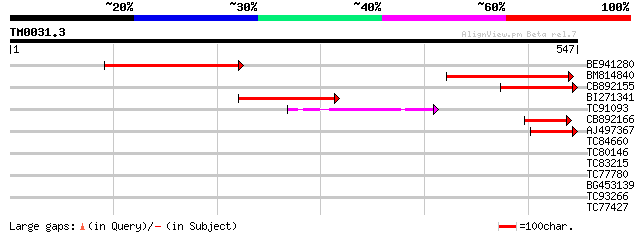
Query= TM0031.3
(547 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BE941280 weakly similar to GP|20197614|g unknown protein {Arabid... 169 3e-42
BM814840 weakly similar to GP|15128241|db helicase-like protein ... 135 5e-32
CB892155 similar to PIR|D86481|D86 hypothetical protein AAG28292... 86 4e-17
BI271341 75 6e-14
TC91093 weakly similar to PIR|T01873|T01873 hypothetical protein... 72 7e-13
CB892166 similar to GP|20197614|gb unknown protein {Arabidopsis ... 45 5e-05
AJ497367 similar to GP|14140286|gb putative helicase {Oryza sati... 45 9e-05
TC84660 similar to PIR|D86481|D86481 hypothetical protein AAG282... 39 0.007
TC80146 39 0.007
TC83215 weakly similar to PIR|F96713|F96713 unknown protein T6L1... 29 4.1
TC77780 weakly similar to GP|23237921|dbj|BAC16494. contains EST... 29 5.3
BG453139 similar to GP|5817349|gb| putative NBS-LRR type disease... 29 5.3
TC93266 similar to PIR|T45709|T45709 hypothetical protein F1P2.4... 28 9.1
TC77427 similar to GP|16648995|gb|AAL24349.1 putative protein {A... 28 9.1
>BE941280 weakly similar to GP|20197614|g unknown protein {Arabidopsis
thaliana}, partial (8%)
Length = 403
Score = 169 bits (428), Expect = 3e-42
Identities = 86/134 (64%), Positives = 98/134 (72%)
Frame = -1
Query: 92 LYGFGGTGKTFVWNILYAALRSRGLIVLNVASSGIASLLLPGDRTAHSRFSIPISINEIS 151
LY +GGT KTF+W L AALRS G IVL ASSGI +LL+PG RTAHSRF IP I+E S
Sbjct: 403 LYDYGGTEKTFIWRALSAALRSEGEIVLACASSGIDALLMPGGRTAHSRFGIPFIIDETS 224
Query: 152 TCNLRQGSPKAELLKKASLIIWDEAPMLNKHCIEALDRSLNDIMKTQSTHGYDIPFGGKV 211
C + P A L+ KA LIIWDEAPM++KHC EALDRSL D++KT DIPFGGKV
Sbjct: 223 MCGVTPNIPLASLVIKAKLIIWDEAPMMHKHCFEALDRSLRDVLKTVDERNKDIPFGGKV 44
Query: 212 VVLGDDFGQILPVI 225
VVLG DF QIL V+
Sbjct: 43 VVLGGDFRQILLVM 2
>BM814840 weakly similar to GP|15128241|db helicase-like protein {Oryza
sativa (japonica cultivar-group)}, partial (6%)
Length = 733
Score = 135 bits (339), Expect = 5e-32
Identities = 69/123 (56%), Positives = 90/123 (73%)
Frame = +3
Query: 422 NGTRMIVNALTKYIIVVTVLNENKMGETTFIPRMSLTPSNSDIPFKFQRRQFSVTLCFVM 481
+GTR+I+ +L K +I V+ GE ++IPRM+L PS +++ F+R QF + L F M
Sbjct: 27 HGTRLIIVSLGKNVICARVIGGTHAGEVSYIPRMNLIPSGANVSITFERCQFPLVLSFAM 206
Query: 482 TINKSQGQSLSHVGLYLPRPVFTHGQPYVALYRVKSRKMLKMLIIDDE*VVSNTTRNVVY 541
TINKSQGQ+L+ VGLYLPRPVFTHGQ YVA+ RVKSR LK+LI D+ S++T NVVY
Sbjct: 207 TINKSQGQTLTSVGLYLPRPVFTHGQLYVAVSRVKSRSGLKILITDENGSPSSSTVNVVY 386
Query: 542 QEV 544
QEV
Sbjct: 387 QEV 395
>CB892155 similar to PIR|D86481|D86 hypothetical protein AAG28292.1
[imported] - Arabidopsis thaliana, partial (4%)
Length = 572
Score = 85.9 bits (211), Expect = 4e-17
Identities = 45/74 (60%), Positives = 54/74 (72%)
Frame = -2
Query: 474 SVTLCFVMTINKSQGQSLSHVGLYLPRPVFTHGQPYVALYRVKSRKMLKMLIIDDE*VVS 533
SV + F MTINKSQGQSL H+G+YLP VF+HGQ YVAL RV SR+ LK+LI +D+
Sbjct: 373 SVEVYFAMTINKSQGQSLKHIGVYLPSSVFSHGQLYVALSRVTSREGLKILISNDDGEDD 194
Query: 534 NTTRNVVYQEVLDN 547
T NVVY+EV N
Sbjct: 193 CVTSNVVYREVFHN 152
>BI271341
Length = 468
Score = 75.1 bits (183), Expect = 6e-14
Identities = 40/98 (40%), Positives = 61/98 (61%)
Frame = +1
Query: 221 ILPVISKGSRSEIVGSAINSSYLWKHCKVMKLTVNMSLQNATSTSSATEIKEFVDWLLQV 280
ILPVI +GSRS+I+ + INSS + HC+V++L NM LQ +S+ E ++F +L+V
Sbjct: 10 ILPVIPRGSRSDIIHATINSSCI*DHCQVVRLKKNMWLQQNGQSSNDPEFEQFSK*ILKV 189
Query: 281 GDGTVKTIDEEETLIEIPPDLLIEQCKEPLLELVNFAY 318
GDG + ++ I+IPP+LLI + L +V Y
Sbjct: 190 GDGKIYEPNDSYADIDIPPELLISNYDDSLQTIVQSTY 303
>TC91093 weakly similar to PIR|T01873|T01873 hypothetical protein T24M8.10 -
Arabidopsis thaliana, partial (11%)
Length = 701
Score = 71.6 bits (174), Expect = 7e-13
Identities = 45/145 (31%), Positives = 70/145 (48%)
Frame = +3
Query: 269 EIKEFVDWLLQVGDGTVKTIDEEETLIEIPPDLLIEQCKEPLLELVNFAYPKLAHNLQKS 328
E K F + L G + ++ ++ PP +P+ +V YP L
Sbjct: 306 EFKTFAEILT----GKMSEPNDSYAEVDTPPG-------DPIDAIVQSTYPNLVSQYNNE 452
Query: 329 SFFQERAILAPTLESLEEINNFMLAMIPGDETEYLSCDTPCKSDEDSGDNAE*FTSEVLN 388
F Q RAIL T E +++IN+++L +IPG+E S + +D + D E L
Sbjct: 453 QFLQSRAILTSTDEVVDQINDYVLKLIPGEERVIYSANRSEVNDVQAFDA---IPPEFLQ 623
Query: 389 DFKCSEIPNHAIKLKAGVPIMLIRN 413
K S++PNH + LK G PIML+R+
Sbjct: 624 SLKTSDLPNHKLTLKVGTPIMLLRD 698
>CB892166 similar to GP|20197614|gb unknown protein {Arabidopsis thaliana},
partial (3%)
Length = 748
Score = 45.4 bits (106), Expect = 5e-05
Identities = 23/46 (50%), Positives = 31/46 (67%)
Frame = -1
Query: 497 YLPRPVFTHGQPYVALYRVKSRKMLKMLIIDDE*VVSNTTRNVVYQ 542
+ R VF+HGQ YVA+ RV SR LK+L+ID+ + T NVVY+
Sbjct: 289 FYDREVFSHGQLYVAISRVSSRNGLKILMIDENGDCIDNTTNVVYK 152
>AJ497367 similar to GP|14140286|gb putative helicase {Oryza sativa (japonica
cultivar-group)}, partial (1%)
Length = 543
Score = 44.7 bits (104), Expect = 9e-05
Identities = 23/45 (51%), Positives = 32/45 (71%)
Frame = +1
Query: 503 FTHGQPYVALYRVKSRKMLKMLIIDDE*VVSNTTRNVVYQEVLDN 547
F++G+ YVA+ RV SRK LK+L+ ++ NTT NVVY+EV N
Sbjct: 19 FSNGKLYVAVSRVTSRKGLKILLAHEDGNCMNTTSNVVYKEVFRN 153
>TC84660 similar to PIR|D86481|D86481 hypothetical protein AAG28292.1
[imported] - Arabidopsis thaliana, partial (1%)
Length = 1009
Score = 38.5 bits (88), Expect = 0.007
Identities = 18/30 (60%), Positives = 23/30 (76%)
Frame = +2
Query: 490 SLSHVGLYLPRPVFTHGQPYVALYRVKSRK 519
S++ G+YLP+P+F HG YVAL RV SRK
Sbjct: 68 SMATKGMYLPQPIF*HG*LYVALSRVTSRK 157
>TC80146
Length = 476
Score = 38.5 bits (88), Expect = 0.007
Identities = 16/35 (45%), Positives = 26/35 (73%)
Frame = -3
Query: 475 VTLCFVMTINKSQGQSLSHVGLYLPRPVFTHGQPY 509
+ + + TINKS+ QSLS++ +YL RP+F+H + Y
Sbjct: 471 IQIPYFKTINKSR*QSLSYMKIYLSRPIFSHEEMY 367
>TC83215 weakly similar to PIR|F96713|F96713 unknown protein T6L1.9
[imported] - Arabidopsis thaliana, partial (21%)
Length = 731
Score = 29.3 bits (64), Expect = 4.1
Identities = 34/152 (22%), Positives = 68/152 (44%), Gaps = 19/152 (12%)
Frame = +2
Query: 216 DDFGQILPVISKGSRSEIVGSAINSSYLWKHCKVMKLTVNMSLQNATSTSSATE------ 269
D+ ++L ISK + + N + + +KL + + ++ + +A E
Sbjct: 149 DNATEVLMGISKSIMGRLQVTEFNLNGYMQRENELKLNIQILIEQLKAKDAALEKNGRCN 328
Query: 270 -----IKEFVDWLLQVGDGTVKTIDEEETLIEIPPDLLIEQ---CKEPLLELVNFAYP-- 319
I+E + +L + + +K ++EE+ E+ + L E+ C E L+E+ NFA
Sbjct: 329 VNVDDIQENTE-VLDLRE-KMKILEEEQKNFEVQINSLSEENEACHEQLIEIENFAESLK 502
Query: 320 ---KLAHNLQKSSFFQERAILAPTLESLEEIN 348
+A N +S+ + + LE EE+N
Sbjct: 503 ESIDIAXNRAESAEAKVTQLTETNLELTEELN 598
>TC77780 weakly similar to GP|23237921|dbj|BAC16494. contains ESTs
D41667(S4320) AU033165(S4320)~mucin-like protein,
partial (13%)
Length = 1215
Score = 28.9 bits (63), Expect = 5.3
Identities = 12/38 (31%), Positives = 22/38 (57%), Gaps = 1/38 (2%)
Frame = -1
Query: 28 CLPYPKFSEIHNFEVIFVADELN-YNRAEMVKIHDEFV 64
C +PKF N ++IF+ E N + +M+K ++ F+
Sbjct: 696 CFRHPKFPSFTNHKIIFIFGECNTIGKGQMIKDNNGFI 583
>BG453139 similar to GP|5817349|gb| putative NBS-LRR type disease resistance
protein {Pisum sativum}, partial (65%)
Length = 638
Score = 28.9 bits (63), Expect = 5.3
Identities = 16/52 (30%), Positives = 27/52 (51%), Gaps = 2/52 (3%)
Frame = -2
Query: 59 IHDEFVSSLTSEQEIFYKNVLDDV--LSDNS*FFFLYGFGGTGKTFVWNILY 108
+ F+ S++E +L DV ++DN ++G GG GKT + +LY
Sbjct: 454 VDQSFMVGRKSDKEKLTNMLLSDVSTVNDNINVVSIFGMGGVGKTALAQLLY 299
>TC93266 similar to PIR|T45709|T45709 hypothetical protein F1P2.40 -
Arabidopsis thaliana, partial (51%)
Length = 855
Score = 28.1 bits (61), Expect = 9.1
Identities = 21/63 (33%), Positives = 34/63 (53%)
Frame = +1
Query: 217 DFGQILPVISKGSRSEIVGSAINSSYLWKHCKVMKLTVNMSLQNATSTSSATEIKEFVDW 276
D+ I+P SKG +S + ++C+V++ TVN S N T S + I+E ++
Sbjct: 376 DYDHIVPY-SKGGQSTL-----------ENCQVLQATVNRSKGNRTDISKSDLIQE--EF 513
Query: 277 LLQ 279
LLQ
Sbjct: 514 LLQ 522
>TC77427 similar to GP|16648995|gb|AAL24349.1 putative protein {Arabidopsis
thaliana}, partial (84%)
Length = 1950
Score = 28.1 bits (61), Expect = 9.1
Identities = 20/47 (42%), Positives = 24/47 (50%)
Frame = +3
Query: 433 KYIIVVTVLNENKMGETTFIPRMSLTPSNSDIPFKFQRRQFSVTLCF 479
KYIIV+ +L K P S +PS+S IPF Q F TL F
Sbjct: 1284 KYIIVLQLLL*KKKKIYPLFPSPSPSPSHSSIPFLLQ--VF*FTLAF 1418
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.139 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,969,957
Number of Sequences: 36976
Number of extensions: 210334
Number of successful extensions: 924
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 920
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 923
length of query: 547
length of database: 9,014,727
effective HSP length: 101
effective length of query: 446
effective length of database: 5,280,151
effective search space: 2354947346
effective search space used: 2354947346
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0031.3


