

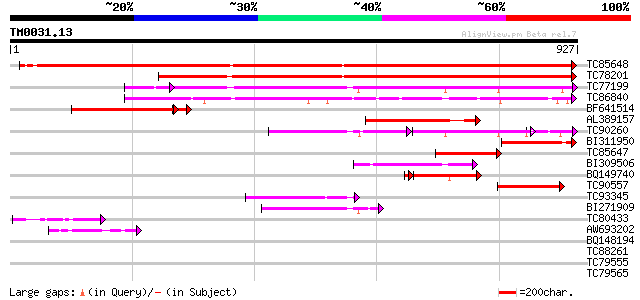
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0031.13
(927 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85648 similar to GP|20197419|gb|AAC77862.2 Argonaute (AGO1)-li... 1555 0.0
TC78201 similar to GP|20197419|gb|AAC77862.2 Argonaute (AGO1)-li... 1147 0.0
TC77199 similar to SP|O04379|AGO1_ARATH Argonaute protein. [Mous... 432 e-127
TC86840 similar to PIR|H86438|H86438 protein T19E23.7 [imported]... 311 1e-84
BF641514 weakly similar to GP|20197419|gb Argonaute (AGO1)-like ... 291 7e-83
AL389157 similar to GP|6539559|dbj ESTs AU068544(C30430) C98487(... 282 4e-76
TC90260 homologue to SP|Q9XGW1|PINH_ARATH PINHEAD protein (ZWILL... 175 1e-71
BI311950 similar to GP|20197419|gb| Argonaute (AGO1)-like protei... 207 1e-53
TC85647 similar to GP|6539559|dbj|BAA88176.1 ESTs AU068544(C3043... 202 5e-52
BI309506 similar to GP|12597784|gb pinhead-like protein {Arabido... 110 3e-24
BQ149740 similar to GP|21280323|db AGO1 homologous protein {Oryz... 107 3e-23
TC90557 similar to SP|O04379|AGO1_ARATH Argonaute protein. [Mous... 105 8e-23
TC93345 similar to SP|Q9SJK3|AGOL_ARATH Argonaute-like protein A... 95 1e-19
BI271909 similar to SP|Q9XGW1|PINH PINHEAD protein (ZWILLE prote... 91 2e-18
TC80433 similar to GP|8778710|gb|AAF79718.1| T1N15.2 {Arabidopsi... 57 4e-08
AW693202 similar to GP|12597784|gb| pinhead-like protein {Arabid... 49 9e-06
BQ148194 homologue to GP|17064856|gb| Unknown protein {Arabidops... 35 0.13
TC88261 similar to PIR|S22697|S22697 extensin - Volvox carteri (... 34 0.23
TC79555 homologue to GP|10567859|gb|AAG18593.1 L8019.5 {Leishman... 33 0.39
TC79565 similar to SP|Q9ZQ70|WRK3_ARATH Probable WRKY transcript... 33 0.66
>TC85648 similar to GP|20197419|gb|AAC77862.2 Argonaute (AGO1)-like protein
{Arabidopsis thaliana}, partial (90%)
Length = 3118
Score = 1555 bits (4025), Expect = 0.0
Identities = 761/912 (83%), Positives = 837/912 (91%), Gaps = 1/912 (0%)
Frame = +3
Query: 16 NEDLMPPPPPPPIVPADVEPVKVDLLDLPPEPVKKKLPTRLPIARKGLGSKGTKLPLLTN 75
NE+ +PPPPP IVPAD+EP+K++ P+ VKKKLPT++P+AR+GLGSKG KLPLLTN
Sbjct: 90 NEECLPPPPP--IVPADIEPIKIE-----PQIVKKKLPTKVPMARRGLGSKGAKLPLLTN 248
Query: 76 HFKVTVANSDGHFFQYSVALSYEDGRPVEGKGVGRKVIDKVQETYGSELNGKDFAYDGEK 135
HFKV V N+DG+FFQYSVAL YEDGRPVEGKG GRK++D+VQETYGSELNGKD AYDGEK
Sbjct: 249 HFKVNVTNTDGYFFQYSVALFYEDGRPVEGKGAGRKILDRVQETYGSELNGKDLAYDGEK 428
Query: 136 TLFTIGSLARNKLEFTVVLEDVISNRNNGNCSPDG-ASTNDSDKKRMRRPYHSKTFKVEI 194
TLFTIGSLA+NKLEFTVVLEDV SNRNNGN SPDG S ND+D+KR+++ + SKT+KVEI
Sbjct: 429 TLFTIGSLAQNKLEFTVVLEDVTSNRNNGNASPDGHGSPNDTDRKRLKKSHRSKTYKVEI 608
Query: 195 SFAAKIPLQAIVNALRGQESENYQEAIRVLDIILRQHAAKQGCLLVRQSFFHNDPKNYAD 254
SFA+KIPLQAI NAL+G E+ENYQEAIRVLDIILRQHAAKQGCLLVRQ+FFHNDPKN+ D
Sbjct: 609 SFASKIPLQAIANALKGHETENYQEAIRVLDIILRQHAAKQGCLLVRQNFFHNDPKNFTD 788
Query: 255 VGGGVLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQPGPVVDFLIANQNVRDPFSLDWAKA 314
VGGGVLGCRG HSSFRTTQSGLSLNIDVSTTMI+ PGPVVDFLIANQNVRDPFSLDW KA
Sbjct: 789 VGGGVLGCRGLHSSFRTTQSGLSLNIDVSTTMIVHPGPVVDFLIANQNVRDPFSLDWNKA 968
Query: 315 KRTLKNLRIKASPSNQEYKITGLSELPCKEQTFTMKKKGGNNGEEDATEEEITVYEYFVN 374
KRTLKNLRI SP+NQEYKITGLSE+PCK+Q FT+KK+G GE+D EEITVY+YFVN
Sbjct: 969 KRTLKNLRITTSPTNQEYKITGLSEMPCKDQLFTLKKRGAVPGEDDT--EEITVYDYFVN 1142
Query: 375 YRKIDLRYSADLPCINVGKPKRPTYVPVELCSLVSLQRYTKALTTLQRSSLVEKSRQKPL 434
RKI L+YSADLPCINVGKPKRPT+VPVELCSLVSLQRYTKAL+TLQRSSLVEKSRQKP
Sbjct: 1143RRKISLQYSADLPCINVGKPKRPTFVPVELCSLVSLQRYTKALSTLQRSSLVEKSRQKPQ 1322
Query: 435 ERMNVLNQALKTSNYGNEPMLKNCGITIASGFTQVEGRVLQAPRLKFGNGEDFNPRNGRW 494
ERM VL ALKTS+YG+EPML+NCGI+I SGFTQV+GRVLQAPRLKFGNGEDFNPRNGRW
Sbjct: 1323ERMRVLTDALKTSDYGSEPMLRNCGISITSGFTQVDGRVLQAPRLKFGNGEDFNPRNGRW 1502
Query: 495 NLNNKKVVRPAKIEHWAVVNFSARCDVRGLVRDLIKCARLKGIPIDEPYEEIFEENGQFR 554
N NNKK+V+P KIE WAVVNFSARCDVRGLVRDLIKC +KGI +++P++ FEENGQFR
Sbjct: 1503NFNNKKIVQPVKIEKWAVVNFSARCDVRGLVRDLIKCGGMKGIHVEQPFD-CFEENGQFR 1679
Query: 555 RAPPLVRVEKMFERIQKELPGAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCISPT 614
RAPPLVRVEKMFE +Q +LPGAP FLLCLL ERKNSDLYGPWKKKNLAE+GIVTQCI+PT
Sbjct: 1680RAPPLVRVEKMFEHVQSKLPGAPKFLLCLLSERKNSDLYGPWKKKNLAEFGIVTQCIAPT 1859
Query: 615 RVNDQYLTNVLMKINAKLGGLNSVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPS 674
RVNDQYLTNVL+KINAKLGG+NS+LGVE +PSIPIVSK PT+ILGMDVSHGSPGQ++IPS
Sbjct: 1860RVNDQYLTNVLLKINAKLGGMNSLLGVEHSPSIPIVSKAPTLILGMDVSHGSPGQTEIPS 2039
Query: 675 IAAVVSSREWPLISKYRACVRTQSPKVEMIDNLFKQVSEKEDEGIIRELLIDFYSSSGKR 734
IAAVVSSR+WPLISKYRACVRTQ KVEMIDNLFK VS+ EDEGIIRELLIDFY+SSG R
Sbjct: 2040IAAVVSSRQWPLISKYRACVRTQGAKVEMIDNLFKPVSDTEDEGIIRELLIDFYNSSGNR 2219
Query: 735 KPDNIIIFRDGVSESQFNQVLNIELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFFQPG 794
KPDNIIIFRDGVSESQFNQVLNIEL+QIIEACKFLDE WNPKFLVIVAQKNHHTKFFQPG
Sbjct: 2220KPDNIIIFRDGVSESQFNQVLNIELSQIIEACKFLDEKWNPKFLVIVAQKNHHTKFFQPG 2399
Query: 795 SPDNVPPGTVIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHS 854
SPDNVPPGTV+DNKICHPRN DFYMCAHAGMIGTSRPTHYHVLLD+IGFSPD+LQELVHS
Sbjct: 2400SPDNVPPGTVVDNKICHPRNYDFYMCAHAGMIGTSRPTHYHVLLDEIGFSPDDLQELVHS 2579
Query: 855 LSYVYQRSTTAISVVAPICYAHLAATQIGQFMKFEDKSDTSSSHGGLTAAGVAPVVPQLP 914
LSYVYQRSTTAISVVAPICYAHLAA+Q+GQFMKFEDKS+TSSSHGG A +PQLP
Sbjct: 2580LSYVYQRSTTAISVVAPICYAHLAASQVGQFMKFEDKSETSSSHGGSGRDINASPIPQLP 2759
Query: 915 KLQDSVSSSMFF 926
KL DSV +SMFF
Sbjct: 2760KLMDSVCNSMFF 2795
>TC78201 similar to GP|20197419|gb|AAC77862.2 Argonaute (AGO1)-like protein
{Arabidopsis thaliana}, partial (71%)
Length = 2047
Score = 1147 bits (2967), Expect = 0.0
Identities = 555/684 (81%), Positives = 619/684 (90%), Gaps = 1/684 (0%)
Frame = +3
Query: 244 FFHNDPKNYADVGGGVLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQPGPVVDFLIANQNV 303
FFHNDPKN+ADVGGGVLGCRGFHSSFR TQSGLSLNIDVSTTMIIQPGPVVDFLI+NQNV
Sbjct: 3 FFHNDPKNFADVGGGVLGCRGFHSSFRATQSGLSLNIDVSTTMIIQPGPVVDFLISNQNV 182
Query: 304 RDPFSLDWAKAKRTLKNLRIKASPSNQEYKITGLSELPCKEQTFTMKKKGGNNGEEDATE 363
RDPF +DW KAKRTLKNLR+K PSNQE+KI GLSE+PCKE TFT+KK+ G+
Sbjct: 183 RDPFQIDWGKAKRTLKNLRVKTHPSNQEWKICGLSEVPCKELTFTLKKRDGDG------T 344
Query: 364 EEITVYEYFVNYRKIDLRYSADLPCINVGKPKRPTYVPVELCSLVSLQRYTKALTTLQRS 423
+E+TV +YF N RKIDLRYSADLPCINVG+PKRPTY P+ELC LVSLQRYTKAL+TLQR+
Sbjct: 345 DEMTVLDYFTNVRKIDLRYSADLPCINVGRPKRPTYFPIELCELVSLQRYTKALSTLQRA 524
Query: 424 SLVEKSRQKPLERMNVLNQALKTSNYGNEPMLKNCGITIASGFTQVEGRVLQAPRLKFGN 483
SLVEKSRQKP ERM +L+ ALKTSNYG EP+L++CGI+I++GFTQVEGRVL AP+LKFGN
Sbjct: 525 SLVEKSRQKPQERMRILSDALKTSNYGAEPLLQSCGISISTGFTQVEGRVLPAPKLKFGN 704
Query: 484 GEDFNPRNGRWNLNNKKVVRPAK-IEHWAVVNFSARCDVRGLVRDLIKCARLKGIPIDEP 542
GEDF PRNGRWN NNKK V+P K IE WAV NFSARCDVRGLVRD+I+ +KGI ID+P
Sbjct: 705 GEDFTPRNGRWNFNNKKFVQPTKKIEKWAVANFSARCDVRGLVRDIIRIGNMKGIMIDQP 884
Query: 543 YEEIFEENGQFRRAPPLVRVEKMFERIQKELPGAPSFLLCLLPERKNSDLYGPWKKKNLA 602
++ +FEEN QFRRAPP+VRVEKMFE IQ +LPGAP FLLCLLP+RKN D+YGPWKKKNLA
Sbjct: 885 FD-VFEENPQFRRAPPMVRVEKMFEDIQSKLPGAPQFLLCLLPDRKNCDIYGPWKKKNLA 1061
Query: 603 EYGIVTQCISPTRVNDQYLTNVLMKINAKLGGLNSVLGVEMNPSIPIVSKVPTIILGMDV 662
++GIV QC+ P RVND YL N+++KINAKLGGLNS+LGVE +PS+PIVSK PT+ILGMDV
Sbjct: 1062DFGIVNQCMCPLRVNDNYLGNIMLKINAKLGGLNSLLGVESSPSLPIVSKAPTLILGMDV 1241
Query: 663 SHGSPGQSDIPSIAAVVSSREWPLISKYRACVRTQSPKVEMIDNLFKQVSEKEDEGIIRE 722
SHGSPGQ+DIPSIAAVVSSR+WPLISKYRACVRTQS KVEMIDNLFK+VS+ EDEGI+RE
Sbjct: 1242SHGSPGQTDIPSIAAVVSSRQWPLISKYRACVRTQSAKVEMIDNLFKKVSDTEDEGIMRE 1421
Query: 723 LLIDFYSSSGKRKPDNIIIFRDGVSESQFNQVLNIELNQIIEACKFLDETWNPKFLVIVA 782
LL+DFY+SS RKPDNIIIFRDGVSESQFNQVLNIEL+QIIEACKFLDE W PKF+VIVA
Sbjct: 1422LLLDFYTSSKNRKPDNIIIFRDGVSESQFNQVLNIELDQIIEACKFLDENWTPKFVVIVA 1601
Query: 783 QKNHHTKFFQPGSPDNVPPGTVIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIG 842
QKNHHT+FFQP SPDNVPPGTVIDNKICHPRN DFY+CAHAGMIGTSRPTHYHVLLD+IG
Sbjct: 1602QKNHHTRFFQPNSPDNVPPGTVIDNKICHPRNYDFYLCAHAGMIGTSRPTHYHVLLDEIG 1781
Query: 843 FSPDELQELVHSLSYVYQRSTTAISVVAPICYAHLAATQIGQFMKFEDKSDTSSSHGGLT 902
FSPDELQEL HSLSYVYQRSTTAISVVAPICYAHLAATQ+GQFMKFEDKS+TSSSHGGL+
Sbjct: 1782FSPDELQELGHSLSYVYQRSTTAISVVAPICYAHLAATQLGQFMKFEDKSETSSSHGGLS 1961
Query: 903 AAGVAPVVPQLPKLQDSVSSSMFF 926
AAG P VP LPKLQD+V + MFF
Sbjct: 1962AAGAVP-VPXLPKLQDNVCNXMFF 2030
>TC77199 similar to SP|O04379|AGO1_ARATH Argonaute protein. [Mouse-ear
cress] {Arabidopsis thaliana}, partial (74%)
Length = 2714
Score = 432 bits (1112), Expect(2) = e-127
Identities = 267/712 (37%), Positives = 394/712 (54%), Gaps = 48/712 (6%)
Frame = +2
Query: 264 GFHSSFRTTQSGLSLNIDVSTTMIIQPGPVVDFL--IANQNV--RDPFSLDWAKAKRTLK 319
GF+ S R TQ GLSLNID+S+T I+P PV++F+ + N++V R D K K+ L+
Sbjct: 293 GFYQSIRPTQMGLSLNIDMSSTAFIEPLPVIEFVTQLLNRDVSARPLSDADRVKIKKALR 472
Query: 320 NLRIKASPSN---QEYKITGLSELPCKEQTFTMKKKGGNNGEEDATEEEITVYEYFVNYR 376
++++ + ++Y+I GL+ +E TF + ++G +V EYF
Sbjct: 473 GIKVEVTHRGNMRRKYRIAGLTSQATRELTFPVDERGTMK----------SVVEYFFETY 622
Query: 377 KIDLRYSADLPCINVGKPKRPTYVPVELCSLVSLQRYTKALTTLQRSSLVEKSRQKPLER 436
++++ PC+ VG P+RP Y+P+E+C +V QRY+K L Q ++L++ + Q+PL+R
Sbjct: 623 GFVIQHT-QWPCLQVGNPQRPNYLPMEVCKIVEGQRYSKRLNERQITALLKVTCQRPLDR 799
Query: 437 MNVLNQALKTSNYGNEPMLKNCGITIASGFTQVEGRVLQAPRLKF---GNGEDFNPRNGR 493
+ Q + + Y +P K GI I+ QVE R+L AP LK+ G +D P+ G+
Sbjct: 800 ERDIMQTVHHNAYHEDPYAKEFGIKISDRLAQVEARILPAPWLKYNDTGREKDCLPQVGQ 979
Query: 494 WNLNNKKVVRPAKIEHWAVVNFSARCD---VRGLVRDLIKCARLKGIPID-EPYEEIFEE 549
WN+ NKK+ +++W VNFS RG +L + + G+ + EP
Sbjct: 980 WNMMNKKMFNGGSVKYWLCVNFSRTVQDSVARGFCYELAQMCYVSGMEFNAEPVVPALTA 1159
Query: 550 NGQFRRAPPLVRVEKMFER----IQKELPGAPSF--LLCLLPERKNSDLYGPWKKKNLAE 603
+VEK+ + + ++P L+ +LP+ N LYG K+ +
Sbjct: 1160RPD--------QVEKVLKNRYHDAKSKMPKDKELDLLIVILPDN-NGSLYGDLKRICETD 1312
Query: 604 YGIVTQCISPTRV---NDQYLTNVLMKINAKLGGLNSVLGVEMNPSIPIVSKVPTIILGM 660
G+V+QC V + QYL NV +KIN K+GG N+VL ++ IP+VS PTII G
Sbjct: 1313LGVVSQCCLTKHVFKMSKQYLANVALKINVKVGGRNTVLVDALSRRIPLVSDRPTIIFGA 1492
Query: 661 DVSHGSPGQSDIPSIAAVVSSREWPLISKYRACVRTQSPKVEMIDNLFKQ----VSEKED 716
DV+H PG+ PSIAAVV+S++WP I+KY V Q+ + E+I +LFKQ V
Sbjct: 1493DVTHPHPGEDSSPSIAAVVASQDWPEITKYAGLVCAQAHRQELIQDLFKQWQDPVRGTLT 1672
Query: 717 EGIIRELLIDFYSSSGKRKPDNIIIFRDGVSESQFNQVLNIELNQIIEACKFLDETWNPK 776
G+I+ELLI F ++G+ KP II +RDGVSE QF QVL EL+ I +AC L+ + P
Sbjct: 1673GGMIKELLISFRRATGQ-KPQRIIFYRDGVSEGQFYQVLLFELDAIRKACASLEPNYQPP 1849
Query: 777 FLVIVAQKNHHTKFFQPGSPD--------NVPPGTVIDNKICHPRNNDFYMCAHAGMIGT 828
+V QK HHT+ F D N+ PGTV+D+KICHP DFY+C+HAG+ GT
Sbjct: 1850VTFVVVQKRHHTRLFASNHQDKSSVDRSGNILPGTVVDSKICHPTEFDFYLCSHAGIQGT 2029
Query: 829 SRPTHYHVLLDDIGFSPDELQELVHSLSYVYQRSTTAISVVAPICYAHLAATQIGQFMKF 888
SRP HYHVL D+ FS D LQ L ++L Y Y R T ++S+V P YAHLAA + +M+
Sbjct: 2030SRPAHYHVLWDENNFSADGLQSLTNNLCYTYARCTRSVSIVPPAYYAHLAAFRARFYMEP 2209
Query: 889 EDKSDTSSSHGGLT-------------AAGVAPVVPQLPKLQDSVSSSMFFC 927
E S + G ++ A G V LP L+D+V MF+C
Sbjct: 2210ETSDSGSMTSGAVSRGGAGAAVGRSTRAPGANAAVRPLPALKDNVKKVMFYC 2365
Score = 42.0 bits (97), Expect(2) = e-127
Identities = 29/85 (34%), Positives = 47/85 (55%), Gaps = 3/85 (3%)
Frame = +1
Query: 188 KTFKVEISFAAKIPLQAIVNALRGQESENYQEAIRVLDIILRQHAAKQGCLLVRQSFFHN 247
+ FKV I AA+ L + L G++++ QEA++VLDI+LR+ + C V +SF+
Sbjct: 67 REFKVVIKLAARADLHHLGLFLEGRQTDAPQEALQVLDIVLRELPTSRYC-PVGRSFYSP 243
Query: 248 DPKNYADVGGGVLGCRGF---HSSF 269
D +G G+ + F HSS+
Sbjct: 244 DLGRRQPLGEGLESWQWFLPKHSSY 318
>TC86840 similar to PIR|H86438|H86438 protein T19E23.7 [imported] -
Arabidopsis thaliana, partial (34%)
Length = 2940
Score = 311 bits (796), Expect = 1e-84
Identities = 238/782 (30%), Positives = 384/782 (48%), Gaps = 44/782 (5%)
Frame = +3
Query: 189 TFKVEISFAAKIPLQAIVNALRGQESENYQEAIRVLDIILRQHAAKQGCLLVRQSFFHND 248
++ V I+ K+ L+ + + L G ++ ++ +D++++++ A++ L R F N
Sbjct: 324 SYTVTITLVNKLELRKLRDYLSGNVYSIPRDILQGMDLVVKENPARRTVSLGRCFFPTNP 503
Query: 249 PKNYADVGGGVLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQPGPVVDFLIANQNVRDPFS 308
P D+ G++ GF S +TT GL+L +D S + V+DFL + ++R
Sbjct: 504 PLIQRDLEPGIIAIGGFQHSLKTTAQGLALCLDYSVLSFRKKMSVLDFL--HDHIRGFNL 677
Query: 309 LDWAKAKR----TLKNLRIKASP--SNQEYKITGLSELPCKEQTFTMKKKGGNNGEEDAT 362
++ K K+ L L++ + + Q+Y I L++ + TF + + G +
Sbjct: 678 AEFRKYKKFVEEVLLGLKVNVTHRRTKQKYTIAKLTDKDTRHITFPILDQEGQTPPRSTS 857
Query: 363 EEEITVYEYFVNYRKIDLRYSADLPCINVGKPKRPTYVPVELCSLVSLQRYTKA-LTTLQ 421
+ YF + D+++ D+P ++ G K +VP+ELC LV QR+ K L
Sbjct: 858 -----LLAYFKDKHNYDIQHK-DIPALDFGGNKT-NFVPMELCVLVEGQRFPKEYLDKNA 1016
Query: 422 RSSLVEKSRQKPLERMNVLNQALKTSNYG-NEPMLKNCGITIASGFTQVEGRVLQAPRLK 480
+L P +R + + +K+S+ +L+N G+ + + T V GRV+ P LK
Sbjct: 1017 AKNLKNMCLASPRDRESTIQMMMKSSDGPCGGGILQNFGMNVNTSMTNVTGRVIGPPMLK 1196
Query: 481 FGNGED------FNPRNGRWNLNNKKVVRPAKIEHWAVVNFSAR----CDVRG--LVRDL 528
G+ +P WNL K +V +E W +++F++ C +RG V +L
Sbjct: 1197 LGDPRGKSTPMKLDPEKCHWNLVGKSMVEGKAVECWGILDFTSDAPNWCKLRGNQFVNNL 1376
Query: 529 IKCARLKGIPIDEPYEEIFE---ENGQFRRAPPLVRVEKMFERIQKELPGAPSFLLCLLP 585
+ R GI ++EP + + G + L+ EK+ E++QK+ FLLC++
Sbjct: 1377 MDKYRKLGIVMNEPVWHEYSAMWKLGDYNLLCELL--EKINEKVQKKCRRRLQFLLCVMA 1550
Query: 586 ERKNSDLYGPWKKKNLAEYGIVTQCISPTRVN---DQYLTNVLMKINAKLGGLNSVLGVE 642
+ W + + GIVTQC N DQYLTN+ +KINAK+GG N VE
Sbjct: 1551 NKDPGYKSLKWIAET--KVGIVTQCCLSGNANEGKDQYLTNLALKINAKIGGSN----VE 1712
Query: 643 MNPSIP-IVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSSREWPLISKYRACVRTQSPKV 701
+ +P + + +G DV+H ++ PSI AVV++ WP ++Y A V Q
Sbjct: 1713 LINRLPHFEDESHVMFIGADVNHPGSRDTNSPSIVAVVATTNWPAANRYAARVCAQEHCT 1892
Query: 702 EMIDNLFKQVSEKEDEGIIRELLIDFYSSSGKRKPDNIIIFRDGVSESQFNQVLNIELNQ 761
E I N G I L+ Y K +P I+IFRDGVSESQF+ VL EL
Sbjct: 1893 EKILNF----------GEICLDLVRHYEKLNKVRPQKIVIFRDGVSESQFHMVLGEELKD 2042
Query: 762 IIEACKFLDETWNPKFLVIVAQKNHHTKFFQPGSPDNVP-----PGTVIDNKICHPRNND 816
+ F + P +IVAQK H T+ F G + P PGTV+D K+ HP D
Sbjct: 2043 LKTV--FQHSNYFPTITLIVAQKRHQTRLFPAGVREGAPSGNVFPGTVVDTKVVHPFEFD 2216
Query: 817 FYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQRSTTAISVVAPICYAH 876
FY+C+H G +GTS+PTHYHVL D+ F+ D LQ+L++ + + + R T +S+V P+ YA
Sbjct: 2217 FYLCSHYGSLGTSKPTHYHVLWDEHRFTSDNLQKLIYDMCFTFARCTKPVSLVPPVYYAD 2396
Query: 877 LAATQIGQFMKFEDKSDT-------SSSHGGLTAAGVAPVV-----PQLPKLQDSVSSSM 924
LAA + G+ + +E K T SSS L ++ ++ P KL + M
Sbjct: 2397 LAAYR-GR-LYYEAKMSTQSPYSTVSSSSSPLASSSISSTASISNDPGFYKLHPDTENGM 2570
Query: 925 FF 926
FF
Sbjct: 2571 FF 2576
>BF641514 weakly similar to GP|20197419|gb Argonaute (AGO1)-like protein
{Arabidopsis thaliana}, partial (17%)
Length = 593
Score = 291 bits (746), Expect(2) = 7e-83
Identities = 137/174 (78%), Positives = 158/174 (90%)
Frame = +2
Query: 102 PVEGKGVGRKVIDKVQETYGSELNGKDFAYDGEKTLFTIGSLARNKLEFTVVLEDVISNR 161
PVEGKGVGRK++D+VQETY S+LNGK+FAYDGEK+LFTIGSL +NKLEF +VLEDV+S+R
Sbjct: 2 PVEGKGVGRKIMDRVQETYASDLNGKEFAYDGEKSLFTIGSLPQNKLEFEIVLEDVVSSR 181
Query: 162 NNGNCSPDGASTNDSDKKRMRRPYHSKTFKVEISFAAKIPLQAIVNALRGQESENYQEAI 221
NNGN SPD N++DKKR+RRPY++KTFKVEISFA K+P+ AI NALRGQE+EN+QEA+
Sbjct: 182 NNGNRSPDANGDNEADKKRVRRPYNAKTFKVEISFATKVPMYAIANALRGQETENFQEAV 361
Query: 222 RVLDIILRQHAAKQGCLLVRQSFFHNDPKNYADVGGGVLGCRGFHSSFRTTQSG 275
RVLDIILRQHAAKQGCLLVRQSFFHNDPKN+ADVGGGVLGC GFH F TQSG
Sbjct: 362 RVLDIILRQHAAKQGCLLVRQSFFHNDPKNFADVGGGVLGCXGFHFKFXATQSG 523
Score = 35.4 bits (80), Expect(2) = 7e-83
Identities = 19/31 (61%), Positives = 21/31 (67%)
Frame = +3
Query: 267 SSFRTTQSGLSLNIDVSTTMIIQPGPVVDFL 297
SS ++GLSLNI V TTMII GPV FL
Sbjct: 498 SSLXLHRAGLSLNIXVXTTMIIHLGPVGGFL 590
>AL389157 similar to GP|6539559|dbj ESTs AU068544(C30430) C98487(E0325)
D23445(C2825) AU068543(C30430) C71774(E0325)
D22458(C11152), partial (18%)
Length = 498
Score = 282 bits (722), Expect = 4e-76
Identities = 145/187 (77%), Positives = 157/187 (83%)
Frame = +3
Query: 583 LLPERKNSDLYGPWKKKNLAEYGIVTQCISPTRVNDQYLTNVLMKINAKLGGLNSVLGVE 642
LL ERKNSDLYGPWKKKNLAE+GIVTQCI+PTR NDQYLTNVL+KINAKLGG+NS+LGVE
Sbjct: 3 LLSERKNSDLYGPWKKKNLAEFGIVTQCIAPTRXNDQYLTNVLLKINAKLGGMNSLLGVE 182
Query: 643 MNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSSREWPLISKYRACVRTQSPKVE 702
+PSIPIVSK PT+ILGMDVSHGSPGQ++IPSIAAVVSSR+WPLISKYRACVRTQ KVE
Sbjct: 183 HSPSIPIVSKAPTLILGMDVSHGSPGQTEIPSIAAVVSSRQWPLISKYRACVRTQGAKVE 362
Query: 703 MIDNLFKQVSEKEDEGIIRELLIDFYSSSGKRKPDNIIIFRDGVSESQFNQVLNIELNQI 762
MIDNLFK VS+ EDEGII RDGVSESQFNQVLNIEL+QI
Sbjct: 363 MIDNLFKPVSDTEDEGII----------------------RDGVSESQFNQVLNIELSQI 476
Query: 763 IEACKFL 769
IEACKFL
Sbjct: 477 IEACKFL 497
>TC90260 homologue to SP|Q9XGW1|PINH_ARATH PINHEAD protein (ZWILLE protein).
[Mouse-ear cress] {Arabidopsis thaliana}, partial (51%)
Length = 2013
Score = 175 bits (444), Expect(2) = 1e-71
Identities = 98/218 (44%), Positives = 131/218 (59%), Gaps = 17/218 (7%)
Frame = +1
Query: 659 GMDVSHGSPGQSDIPSIAAVVSSREWPLISKYRACVRTQSPKVEMIDNLFKQ----VSEK 714
G DV+H G+ PSIAAVV+S++WP ++KY V Q + E+I +L+K V
Sbjct: 748 GADVTHPENGEDSSPSIAAVVASQDWPEVTKYAGLVCAQPQRQELIQDLYKTWHDPVRGV 927
Query: 715 EDEGIIRELLIDFYSSSGKRKPDNIIIFRDGVSESQFNQVLNIELNQIIEACKFLDETWN 774
G+IR+LL+ F ++G+ KP II +RDGVSE QF QVL EL+ I +AC L+ +
Sbjct: 928 VSGGMIRDLLVSFRRATGQ-KPQRIIFYRDGVSEGQFYQVLLYELDAIRKACASLEPNYQ 1104
Query: 775 PKFLVIVAQKNHHTKFFQPGSPD--------NVPPGTVIDNKICHPRNNDFYMCAHAGMI 826
P IV QK HHT+ F D N+ PGTV+D KICHP DFY+C+HAG+
Sbjct: 1105PPVTFIVVQKRHHTRLFANNHKDRNSTDRSGNILPGTVVDTKICHPTEFDFYLCSHAGIQ 1284
Query: 827 GTSRPTHYHVLLDDIGF-----SPDELQELVHSLSYVY 859
GTSRP HYHVL D+ F S + Q L+H + +VY
Sbjct: 1285GTSRPAHYHVLWDENNFTAGWNSVFDKQSLLH-ICHVY 1395
Score = 114 bits (284), Expect(2) = 1e-71
Identities = 78/254 (30%), Positives = 130/254 (50%), Gaps = 19/254 (7%)
Frame = +3
Query: 423 SSLVEKSRQKPLERMNVLNQALKTSNYGNEPMLKNCGITIASGFTQVEGRVLQAPRLKF- 481
+SL++ + Q+P +R N + + ++ + Y +P K GI ++ VE R+L AP LK+
Sbjct: 21 TSLLKVTCQRPRDRENDILRTVQHNAYDQDPYAKEFGIKVSEKLASVEARILPAPWLKYH 200
Query: 482 --GNGEDFNPRNGRWNLNNKKVVRPAKIEHWAVVNFSARCD---VRGLVRDLIKCARLKG 536
G ++ P+ G+WN+ NKK++ + WA +NFS R +L + ++ G
Sbjct: 201 ESGKEKNCLPQVGQWNMMNKKMINGMTVSRWACINFSRGVQDSVARTFCNELAQMCQVSG 380
Query: 537 IPID-EPYEEIFEENGQFRRAPPLVRVEKMFERIQ---------KELPGAPSFLLCLLPE 586
+ + EP I+ + +VEK + + KEL LL +LP+
Sbjct: 381 MEFNLEPVIPIYNAKPE--------QVEKALKHVYHVSSNKTKGKEL----ELLLVILPD 524
Query: 587 RKNSDLYGPWKKKNLAEYGIVTQCISPT---RVNDQYLTNVLMKINAKLGGLNSVLGVEM 643
N LYG K+ + G+++QC ++ QYL NV +KIN K+GG N+VL +
Sbjct: 525 N-NGSLYGDLKRICETDLGLISQCCLTKHVFKITKQYLANVSLKINVKMGGRNTVLVDAV 701
Query: 644 NPSIPIVSKVPTII 657
+ IP+VS +PTII
Sbjct: 702 SCRIPLVSDIPTII 743
Score = 51.6 bits (122), Expect = 1e-06
Identities = 34/86 (39%), Positives = 47/86 (54%), Gaps = 4/86 (4%)
Frame = +2
Query: 846 DELQELVHSLSYVYQRSTTAISVVAPICYAHLAATQIGQFMKFEDKSDTSSSHG----GL 901
D +Q L ++L Y Y T ++SVV P YAHLAA + +F D +T S+ G
Sbjct: 1343 DGIQSLTNNLCYTYATCTRSVSVVPPAYYAHLAAFR-ARFYTEPDLQETGSTGGRGSKTT 1519
Query: 902 TAAGVAPVVPQLPKLQDSVSSSMFFC 927
AAG V P LP L+++V MF+C
Sbjct: 1520 RAAGDCGVKP-LPALKENVKRVMFYC 1594
>BI311950 similar to GP|20197419|gb| Argonaute (AGO1)-like protein
{Arabidopsis thaliana}, partial (10%)
Length = 592
Score = 207 bits (528), Expect = 1e-53
Identities = 99/123 (80%), Positives = 111/123 (89%)
Frame = +3
Query: 804 VIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQRST 863
V+D+KICHPRN DFYMCAHAGMIGTSRPTHYHVLLD+IGFSPD+LQELVHSLSYVYQRST
Sbjct: 3 VVDSKICHPRNYDFYMCAHAGMIGTSRPTHYHVLLDEIGFSPDDLQELVHSLSYVYQRST 182
Query: 864 TAISVVAPICYAHLAATQIGQFMKFEDKSDTSSSHGGLTAAGVAPVVPQLPKLQDSVSSS 923
TAISVVAPICYAHLAA+Q+GQFMKFEDKS+TSSS GG+ A+ ++PQLP L V +S
Sbjct: 183 TAISVVAPICYAHLAASQVGQFMKFEDKSETSSSQGGINAS----LIPQLPNLHKRVCNS 350
Query: 924 MFF 926
MFF
Sbjct: 351 MFF 359
>TC85647 similar to GP|6539559|dbj|BAA88176.1 ESTs AU068544(C30430)
C98487(E0325) D23445(C2825) AU068543(C30430)
C71774(E0325) D22458(C11152), partial (11%)
Length = 1366
Score = 202 bits (514), Expect = 5e-52
Identities = 98/107 (91%), Positives = 101/107 (93%)
Frame = +3
Query: 697 QSPKVEMIDNLFKQVSEKEDEGIIRELLIDFYSSSGKRKPDNIIIFRDGVSESQFNQVLN 756
Q KVEMIDNLFK VS+ EDEGIIRELLIDFY+SSG RKPDNIIIFRDGVSESQFNQVLN
Sbjct: 3 QGAKVEMIDNLFKPVSDTEDEGIIRELLIDFYNSSGNRKPDNIIIFRDGVSESQFNQVLN 182
Query: 757 IELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFFQPGSPDNVPPGT 803
IEL+QIIEACKFLDE WNPKFLVIVAQKNHHTKFFQPGSPDNVPPGT
Sbjct: 183 IELSQIIEACKFLDEKWNPKFLVIVAQKNHHTKFFQPGSPDNVPPGT 323
>BI309506 similar to GP|12597784|gb pinhead-like protein {Arabidopsis
thaliana}, partial (20%)
Length = 737
Score = 110 bits (274), Expect = 3e-24
Identities = 70/209 (33%), Positives = 112/209 (53%), Gaps = 5/209 (2%)
Frame = +1
Query: 562 VEKMFERIQKELPGAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCI---SPTRVND 618
+E +RIQ L+C++ ++ Y K+ G+V+QC + +++
Sbjct: 145 LESKLKRIQSIASNNLQLLICIMEKKHKG--YADLKRIAETSVGVVSQCCLYPNLIKLSS 318
Query: 619 QYLTNVLMKINAKLGGLNSVLGVEMNPSIPIVSKV--PTIILGMDVSHGSPGQSDIPSIA 676
Q+L N+ +KINAK+GG L + +P + + P + +G DV+H P PS+A
Sbjct: 319 QFLANLALKINAKVGGCTVALYNSLPSQLPRLFNIDEPVMFMGADVTHPHPLDDSSPSVA 498
Query: 677 AVVSSREWPLISKYRACVRTQSPKVEMIDNLFKQVSEKEDEGIIRELLIDFYSSSGKRKP 736
AVV S WP +KY + +R+Q+ + E+I +L ++ ELL DFY ++ P
Sbjct: 499 AVVGSMNWPTANKYISRIRSQTHRQEIIADL---------GAMVGELLEDFYQEV-EKLP 648
Query: 737 DNIIIFRDGVSESQFNQVLNIELNQIIEA 765
+ II FRDGVSE+QF +VL EL I +A
Sbjct: 649 NRIIFFRDGVSETQFYKVLQEELQSIKQA 735
>BQ149740 similar to GP|21280323|db AGO1 homologous protein {Oryza sativa
(japonica cultivar-group)}, partial (14%)
Length = 412
Score = 107 bits (266), Expect(2) = 3e-23
Identities = 54/115 (46%), Positives = 76/115 (65%), Gaps = 4/115 (3%)
Frame = +3
Query: 661 DVSHGSPGQSDIPSIAAVVSSREWPLISKYRACVRTQSPKVEMIDNLFKQVSEKEDE--- 717
DV+H PG+ PSIAAVV+S +WP ++KYR V +Q + E+I +L+ + +
Sbjct: 66 DVTHPHPGEDSSPSIAAVVASMDWPCVTKYRGMVSSQIHREEIIQDLYTETKHPQKGTVP 245
Query: 718 -GIIRELLIDFYSSSGKRKPDNIIIFRDGVSESQFNQVLNIELNQIIEACKFLDE 771
G+IRELL+ FY S+ KRKP+ II +RDGVSE QF QVL E++ I +AC +E
Sbjct: 246 GGMIRELLLAFYRSNNKRKPERIIFYRDGVSEGQFXQVLLYEMDAIWKACASFEE 410
Score = 20.8 bits (42), Expect(2) = 3e-23
Identities = 8/14 (57%), Positives = 11/14 (78%)
Frame = +2
Query: 646 SIPIVSKVPTIILG 659
+IP+V+ PTII G
Sbjct: 20 NIPLVTDKPTIIFG 61
>TC90557 similar to SP|O04379|AGO1_ARATH Argonaute protein. [Mouse-ear
cress] {Arabidopsis thaliana}, partial (8%)
Length = 597
Score = 105 bits (262), Expect = 8e-23
Identities = 52/110 (47%), Positives = 71/110 (64%)
Frame = +1
Query: 798 NVPPGTVIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSY 857
N PGTV+++ I HP DFY+ +H G+ GTSRPTHYHVLLD+ GF+ D LQ L ++L Y
Sbjct: 13 NCQPGTVVESVITHPCEFDFYLQSHPGLQGTSRPTHYHVLLDENGFNSDSLQTLSYNLCY 192
Query: 858 VYQRSTTAISVVAPICYAHLAATQIGQFMKFEDKSDTSSSHGGLTAAGVA 907
V+ R T A+S+V P+ YAHL + E+ SD +S G +A+ A
Sbjct: 193 VFARCTRAVSLVPPVYYAHLVCARARFHSSGENWSDPDTSEGAGSASYAA 342
>TC93345 similar to SP|Q9SJK3|AGOL_ARATH Argonaute-like protein At2g27880.
[Mouse-ear cress] {Arabidopsis thaliana}, partial (12%)
Length = 639
Score = 95.1 bits (235), Expect = 1e-19
Identities = 58/190 (30%), Positives = 96/190 (50%), Gaps = 4/190 (2%)
Frame = +3
Query: 386 LPCINVGKPKRPTYVPVELCSLVSLQRYTKALTTLQRSSLVEKSRQKPLERMNVLNQALK 445
LP + G RP ++P+ELC + S QRY+K L Q ++L+ + Q+P +R N + + +
Sbjct: 21 LPALQAGSDTRPMFIPMELCQIESGQRYSKRLNEEQVTNLLRATCQRPQQRENDIKKIVT 200
Query: 446 TSNYGNEPMLKNCGITIASGFTQVEGRVLQAPRLKF-GNGED--FNPRNGRWNLNNKKVV 502
+ + + K GI + V+ RVL P LK+ GNG D PR G+WN+ KVV
Sbjct: 201 QHRFNADKVAKEFGINVREELALVDARVLPPPMLKYHGNGGDSKIEPRMGQWNMIRHKVV 380
Query: 503 RPAKIEHWAVVNFSARCDVRGLVRDLIKCARLKGIPID-EPYEEIFEENGQFRRAPPLVR 561
++ W+ ++FS R + +L+ + KGI + +P I N +
Sbjct: 381 NGGTVQFWSCLSFS-RLNPTQFCAELVNMCQAKGIIFNRDPLVPIAPANSN--------Q 533
Query: 562 VEKMFERIQK 571
+EK E++ K
Sbjct: 534 IEKELEKLDK 563
>BI271909 similar to SP|Q9XGW1|PINH PINHEAD protein (ZWILLE protein).
[Mouse-ear cress] {Arabidopsis thaliana}, partial (23%)
Length = 694
Score = 91.3 bits (225), Expect = 2e-18
Identities = 63/211 (29%), Positives = 101/211 (47%), Gaps = 12/211 (5%)
Frame = +1
Query: 412 RYTKALTTLQRSSLVEKSRQKPLERMNVLNQALKTSNYGNEPMLKNCGITIASGFTQVEG 471
RYTK L Q +SL++ + Q+P +R N + Q ++ + Y +P K GI I+ VE
Sbjct: 1 RYTKRLNEKQITSLLKVTCQRPRDRENDILQTVQHNAYDQDPYAKEFGINISEKLASVEA 180
Query: 472 RVLQAPRLKF---GNGEDFNPRNGRWNLNNKKVVRPAKIEHWAVVNFSARCD---VRGLV 525
R+L P LK+ G ++ P G+WN+ NKK++ + WA +NFS R
Sbjct: 181 RILPTPWLKYHESGKEKNCLPHVGQWNMMNKKMINGMTVNRWACINFSRSVQDSVARTFC 360
Query: 526 RDLIKCARLKGIPID-EPYEEIFEENGQFRRAPPLVRVEKMFERI-----QKELPGAPSF 579
DL + ++ G+ + EP I+ + +VEK + + K
Sbjct: 361 NDLAQMCQVSGMEFNLEPVIPIYNAKPE--------QVEKALKHVYHVSTNKTKGKELEL 516
Query: 580 LLCLLPERKNSDLYGPWKKKNLAEYGIVTQC 610
LL +LP+ N LYG K+ + GI++QC
Sbjct: 517 LLAILPD-SNGSLYGDLKRICETDLGIISQC 606
>TC80433 similar to GP|8778710|gb|AAF79718.1| T1N15.2 {Arabidopsis
thaliana}, partial (13%)
Length = 794
Score = 56.6 bits (135), Expect = 4e-08
Identities = 48/153 (31%), Positives = 63/153 (40%), Gaps = 1/153 (0%)
Frame = +3
Query: 5 EHENGNGNGNGNEDLMPPPPPPPIVPADVEPVKVDLLDLPPEPVKKKLPTRLPIARKGLG 64
E E G D P PPPPP + K R P+ R G G
Sbjct: 414 EVEQDLGQLTIQSDETPAPPPPP---------------------QSKSSLRFPL-RPGKG 527
Query: 65 SKGTKLPLLTNHFKVTVANSDGHFFQYSVALSYEDGRPVEGKGVGRKVIDKVQETYGSEL 124
S G K + NHF + D H QY V ++ E V +GV R V++++ Y
Sbjct: 528 SYGRKTLVKANHFFAELPKKDLH--QYDVTITPE----VTSRGVNRAVMEQLVRLYRDSH 689
Query: 125 NGKDF-AYDGEKTLFTIGSLARNKLEFTVVLED 156
GK AYDG K+L+T G L +F + L D
Sbjct: 690 LGKRLPAYDGRKSLYTAGPLPFISKDFRITLVD 788
>AW693202 similar to GP|12597784|gb| pinhead-like protein {Arabidopsis
thaliana}, partial (7%)
Length = 428
Score = 48.9 bits (115), Expect = 9e-06
Identities = 41/152 (26%), Positives = 65/152 (41%)
Frame = +3
Query: 64 GSKGTKLPLLTNHFKVTVANSDGHFFQYSVALSYEDGRPVEGKGVGRKVIDKVQETYGSE 123
G +G + LL NHF V +S + Y+V ++ P K V R++ K+
Sbjct: 36 GQEGPVISLLANHFLVKF-DSSHKIYHYNVEIT-----PHPSKDVAREIKHKLVNNNAEI 197
Query: 124 LNGKDFAYDGEKTLFTIGSLARNKLEFTVVLEDVISNRNNGNCSPDGASTNDSDKKRMRR 183
L+G AYDG K L++ +KLEF + G P ST+ +K+
Sbjct: 198 LSGALPAYDGRKNLYSPIEFQNDKLEFYI-----------GLPIPTSKSTSPYEKREQH- 341
Query: 184 PYHSKTFKVEISFAAKIPLQAIVNALRGQESE 215
K F++ I +KI + + N L + E
Sbjct: 342 ----KLFRINIKLVSKIDGKGLTNYLSKEGDE 425
>BQ148194 homologue to GP|17064856|gb| Unknown protein {Arabidopsis
thaliana}, partial (8%)
Length = 673
Score = 35.0 bits (79), Expect = 0.13
Identities = 23/68 (33%), Positives = 32/68 (46%), Gaps = 1/68 (1%)
Frame = +2
Query: 19 LMPPPPPPPIVPADVEPVKVDLLDLPPEPVKKKLPTRLPIARKGLGSKG-TKLPLLTNHF 77
L PPPPPPP +P +L+ P+ V +P A++G G +G + P H
Sbjct: 227 LDPPPPPPPPLP---------MLETTPDKVV------IPTAKRGSGRRGRDRRPGSRRHR 361
Query: 78 KVTVANSD 85
KV N D
Sbjct: 362 KVVAGNRD 385
>TC88261 similar to PIR|S22697|S22697 extensin - Volvox carteri (fragment),
partial (7%)
Length = 1516
Score = 34.3 bits (77), Expect = 0.23
Identities = 16/37 (43%), Positives = 19/37 (51%)
Frame = +2
Query: 21 PPPPPPPIVPADVEPVKVDLLDLPPEPVKKKLPTRLP 57
PPPPPPP P+ P PP P+ + LP LP
Sbjct: 371 PPPPPPPSSPSPPPPP-------PPPPILQPLPLSLP 460
>TC79555 homologue to GP|10567859|gb|AAG18593.1 L8019.5 {Leishmania major},
partial (1%)
Length = 1410
Score = 33.5 bits (75), Expect = 0.39
Identities = 22/52 (42%), Positives = 23/52 (43%)
Frame = +2
Query: 19 LMPPPPPPPIVPADVEPVKVDLLDLPPEPVKKKLPTRLPIARKGLGSKGTKL 70
L PPPPPP PA P L PP P P RLP AR+ G L
Sbjct: 656 LSSPPPPPPFFPAR-RPGTRSLPKSPPRP----RPARLPSARRASPRGGRPL 796
>TC79565 similar to SP|Q9ZQ70|WRK3_ARATH Probable WRKY transcription factor
3 (WRKY DNA-binding protein 3). [Mouse-ear cress],
partial (35%)
Length = 1602
Score = 32.7 bits (73), Expect = 0.66
Identities = 15/45 (33%), Positives = 23/45 (50%)
Frame = -1
Query: 12 NGNGNEDLMPPPPPPPIVPADVEPVKVDLLDLPPEPVKKKLPTRL 56
NG+ +PPPPPPP ++V P + DL KL +++
Sbjct: 435 NGDKPPPPLPPPPPPPCSTSNVSPARSSEKDLQSSLSPNKLESKV 301
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,516,752
Number of Sequences: 36976
Number of extensions: 440122
Number of successful extensions: 6562
Number of sequences better than 10.0: 93
Number of HSP's better than 10.0 without gapping: 3296
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4754
length of query: 927
length of database: 9,014,727
effective HSP length: 105
effective length of query: 822
effective length of database: 5,132,247
effective search space: 4218707034
effective search space used: 4218707034
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0031.13


