

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0030.10
(482 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
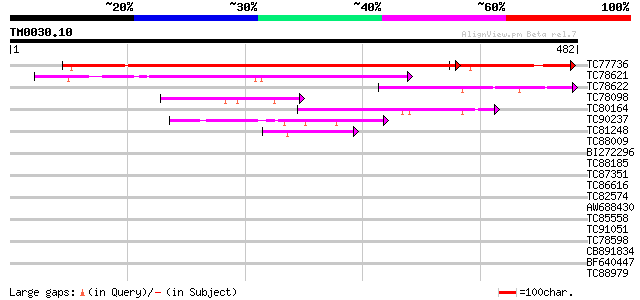
Score E
Sequences producing significant alignments: (bits) Value
TC77736 similar to GP|12324651|gb|AAG52287.1 putative GTP-bindin... 592 0.0
TC78621 similar to GP|6729012|gb|AAF27009.1| putative GTPase {Ar... 152 3e-37
TC78622 weakly similar to GP|14587220|dbj|BAB61154. putative GTP... 74 1e-13
TC78098 similar to PIR|A84670|A84670 probable nucleotide-binding... 71 8e-13
TC80164 similar to PIR|A84670|A84670 probable nucleotide-binding... 63 2e-10
TC90237 similar to GP|20466544|gb|AAM20589.1 unknown protein {Ar... 45 6e-05
TC81248 similar to SP|O81004|YB87_ARATH Hypothetical GTP-binding... 44 2e-04
TC88009 similar to PIR|T48507|T48507 probable GTP-binding protei... 39 0.004
BI272296 similar to GP|12322014|gb GTPase putative; 34281-30152... 35 0.064
TC88185 homologue to PIR|T09368|T09368 GTP-binding protein F23K1... 35 0.083
TC87351 similar to GP|10176988|dbj|BAB10220. GTP-binding protein... 34 0.11
TC86616 similar to GP|7643796|gb|AAF65513.1| GTP-binding protein... 31 1.2
TC82574 similar to GP|17473914|gb|AAL38371.1 GTP-binding protein... 31 1.2
AW688430 similar to GP|22535581|dbj contains EST AU182189(C63973... 30 2.1
TC85558 homologue to GP|7453538|gb|AAF62870.1| Toc64 {Pisum sati... 30 2.1
TC91051 similar to PIR|H84787|H84787 probable receptor-like prot... 30 2.7
TC78598 similar to SP|Q43207|FKB7_WHEAT 70 kDa peptidylprolyl is... 29 4.6
CB891834 homologue to PIR|C96751|C96 probable GTP-binding protei... 28 6.0
BF640447 similar to GP|20152085|gb| RE21802p {Drosophila melanog... 28 6.0
TC88979 similar to PIR|H96601|H96601 hypothetical protein T6H22.... 28 6.0
>TC77736 similar to GP|12324651|gb|AAG52287.1 putative GTP-binding protein;
106556-109264 {Arabidopsis thaliana}, partial (76%)
Length = 2145
Score = 592 bits (1525), Expect(2) = 0.0
Identities = 294/342 (85%), Positives = 314/342 (90%), Gaps = 4/342 (1%)
Frame = +3
Query: 46 QSNSRV----QRPFTPVFGPKTKRKRPCLLAFDYQSLAKKAHVAQEAFEEKYAAGASGTA 101
Q SRV + PF+ FGPKTKRKRP LLA DY+SLAKKA +Q+AFEEK+A+G + A
Sbjct: 420 QKQSRVHLLDREPFSDAFGPKTKRKRPSLLASDYESLAKKADGSQDAFEEKHASGTA--A 593
Query: 102 EANEADGFIDLLGHTMFQKGQTKHISGELYKVIDSSDVVVQVLDARDPQGTRCYHLEKHL 161
EA+EADGF DL+ HTMF+KGQ+K I GELYKVIDSSDVVVQV+DARDPQGTRCYHLEKHL
Sbjct: 594 EADEADGFRDLVRHTMFEKGQSKRIWGELYKVIDSSDVVVQVIDARDPQGTRCYHLEKHL 773
Query: 162 KENCKYKHMMLLLNKCDLIPAWATKGWLRVLSKEYPTLAFHASINKSFGKGSLLSVLRQF 221
K+NCK+KHM+LLLNKCDLIPAWATKGWLRVLSKEYPTLAFHASINKSFGKGSLLSVLRQF
Sbjct: 774 KKNCKHKHMVLLLNKCDLIPAWATKGWLRVLSKEYPTLAFHASINKSFGKGSLLSVLRQF 953
Query: 222 ARLKSDKQAISVGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFLID 281
ARLKSDKQAISVGF+GYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFLID
Sbjct: 954 ARLKSDKQAISVGFVGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFLID 1133
Query: 282 CPGVVYQNKDSEADVVLKGVVRVTNLKDAADRIGEVLKRVMKEHLHRAYKIKEWENENDF 341
CPGVVY N DSE DVVLKGVVRVTNLKDAAD IGEVLKRV KEHL RAYKIKEW +ENDF
Sbjct: 1134CPGVVYHNNDSETDVVLKGVVRVTNLKDAADHIGEVLKRVKKEHLTRAYKIKEWVDENDF 1313
Query: 342 LLQLCKLTGKLLKGGEPDLMTAAKMVLHDWQRGKIPFFVPPP 383
LLQLCK +GKLLKGGEPDLMTAAKM+LHDWQRGKIPFFVPPP
Sbjct: 1314LLQLCKSSGKLLKGGEPDLMTAAKMILHDWQRGKIPFFVPPP 1439
Score = 115 bits (287), Expect(2) = 0.0
Identities = 70/111 (63%), Positives = 78/111 (70%), Gaps = 4/111 (3%)
Frame = +1
Query: 375 KIPFFVPPPQQEDLSE--EPIVNGLDVDDGVDSNQASAAIKAIANVLSSQQQSSVPVQKD 432
K PF +Q+ LSE EP VNG+DVD+ VD NQASAAIKAIANVLSSQQQ +VPVQ D
Sbjct: 1414 KFPFLSLLRRQDALSESEEPNVNGIDVDEAVDHNQASAAIKAIANVLSSQQQRNVPVQGD 1593
Query: 433 LFTENELEEE-ADNLLLISGDSPD-GDVLDSDSDTSEQDPDTEVPYEQPHS 481
L+ ENEL+EE AD L PD GD D DSD SE+DP TEVP EQ S
Sbjct: 1594 LYNENELDEETADQL-------PDTGDYTDEDSDASEEDPSTEVPSEQDPS 1725
Score = 37.4 bits (85), Expect = 0.013
Identities = 24/67 (35%), Positives = 35/67 (51%), Gaps = 4/67 (5%)
Frame = +3
Query: 15 LSFSAMVKKKES----SGKSKQSLDANGNNDAKMEQSNSRVQRPFTPVFGPKTKRKRPCL 70
L S M KKKE SGK K S DAN +ND+K E+ ++ R + ++ +
Sbjct: 45 LRISTMTKKKERNVNLSGKPKHSNDANRSNDSKTEKRSAATVRRLKMYKTRPVRNRKGHV 224
Query: 71 LAFDYQS 77
L+ +YQS
Sbjct: 225 LSNEYQS 245
>TC78621 similar to GP|6729012|gb|AAF27009.1| putative GTPase {Arabidopsis
thaliana}, partial (56%)
Length = 1154
Score = 152 bits (384), Expect = 3e-37
Identities = 106/348 (30%), Positives = 174/348 (49%), Gaps = 27/348 (7%)
Frame = +3
Query: 22 KKKESSGKSKQSLDANGNNDAKMEQSN-----SRVQRPFTPVFGPKTKRKRPCLLAFDYQ 76
KK + SGK K D ND ++ +R +R + K +RK
Sbjct: 153 KKLQLSGKKKVEKDPGIPNDWPFKEQELKLLEARRERALAELAKKKDERKER-------- 308
Query: 77 SLAKKAHVAQEAFEEKYAAGASGTAEANEADGFIDLLGHTMFQKGQTKHISGELYKVIDS 136
A+K ++ + + + E E+ + + T + + +G+L VI +
Sbjct: 309 --ARKRKAGLPVDDDNSNSVETASIENTES---VPTVAKTN-KDSSDRAFAGDLEAVIAT 470
Query: 137 SDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMMLLLNKCDLIPAWATKGWLRVLSKEY 196
SDV+++VLDARDP GTRC ++EK ++++ YK +LLLNK DL+P + + WL+ L +E+
Sbjct: 471 SDVILEVLDARDPLGTRCVNIEKRVRDSGTYKRHVLLLNKIDLVPRESVEKWLKYLREEF 650
Query: 197 PTLAFHASINK---SFGKG------------------SLLSVLRQFARLKSDKQAISVGF 235
PT+AF S + + G+ +LL +L+ +AR K +I+VG
Sbjct: 651 PTVAFKCSTQQQKSNLGRSKKIKTSDTLQLSDWLRAETLLKLLKNYARSDKLKTSITVGL 830
Query: 236 IGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFLIDCPGVV-YQNKDSEA 294
+G PNVG SS+IN++ + V+ G T+ Q I L K + L+DCPGVV +++++A
Sbjct: 831 VGLPNVGNSSLINSVMRSHAVHVSASAGSTRSKQEIRLDKNVKLLDCPGVVMLDSRENDA 1010
Query: 295 DVVLKGVVRVTNLKDAADRIGEVLKRVMKEHLHRAYKIKEWENENDFL 342
+ LK R L D E+L L YKI +++++DFL
Sbjct: 1011TIALKNCKRNEKLADPVSPEKEILMLCPARVLISLYKISSFDSDDDFL 1154
>TC78622 weakly similar to GP|14587220|dbj|BAB61154. putative GTPase {Oryza
sativa (japonica cultivar-group)}, partial (32%)
Length = 1025
Score = 74.3 bits (181), Expect = 1e-13
Identities = 56/174 (32%), Positives = 86/174 (49%), Gaps = 5/174 (2%)
Frame = +1
Query: 314 IGEVLKRVMKEHLHRAYKIKEWENENDFLLQLCKLTGKLLKGGEPDLMTAAKMVLHDWQR 373
+ E+LK L YKI +++ +DFL + + GKL KGG D+ AA++VLHDW
Sbjct: 7 VKEILKLCPARVLISLYKIPSFDSVDDFLQIVATVRGKLKKGGIVDIDAAARIVLHDWNI 186
Query: 374 GKIPFFVPPP--QQEDLSEEPIVNGLDVDDGVDSNQASAAIKAIANVLSSQQQSSVPVQK 431
GKIP++ PP Q + SE IV+ + +D S I N+ + + V V
Sbjct: 187 GKIPYYTMPPVRDQGEPSEAKIVSQFSKEFNIDEVYNSET-SFIGNLKPTVELDHVEVPS 363
Query: 432 D---LFTENELEEEADNLLLISGDSPDGDVLDSDSDTSEQDPDTEVPYEQPHSA 482
+F E LE++ L + P+ +V D+D D S + +V E+ SA
Sbjct: 364 SCPLIFDETMLEDDKPVELAKQDEGPE-NVDDNDEDESMECGKEDVSKEKVKSA 522
>TC78098 similar to PIR|A84670|A84670 probable nucleotide-binding protein
[imported] - Arabidopsis thaliana, partial (40%)
Length = 1102
Score = 71.2 bits (173), Expect = 8e-13
Identities = 53/168 (31%), Positives = 80/168 (47%), Gaps = 46/168 (27%)
Frame = +3
Query: 129 ELYKVIDSSDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMMLLLNKCDLIPA------ 182
+L++V++ SD++V V+D+RDP RC LE + KE +K+ +LL+NK DL+PA
Sbjct: 591 QLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVREKW 770
Query: 183 -------------WATKGWLRVL------SKEYPTLAFHASIN-KSFGKGSLLSVLRQFA 222
W+ K VL S + +A + + K +G+ LL+ L+ A
Sbjct: 771 AEYFRAHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQSEA 950
Query: 223 R--------------------LKSDKQAISVGFIGYPNVGKSSVINTL 250
+ S + VGF+GYPNVGKSS IN L
Sbjct: 951 EAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINAL 1094
>TC80164 similar to PIR|A84670|A84670 probable nucleotide-binding protein
[imported] - Arabidopsis thaliana, partial (31%)
Length = 1215
Score = 63.2 bits (152), Expect = 2e-10
Identities = 49/191 (25%), Positives = 85/191 (43%), Gaps = 19/191 (9%)
Frame = +3
Query: 245 SVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFLIDCPGVVYQN-KDSEADVVLKGVVR 303
S IN L + V P +TK +Q + +++++ L DCPG+V+ + S +++ GV
Sbjct: 3 STINALVGQKKTGVTSTPRKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMIT*GVSP 182
Query: 304 VTNLKDAADRIGEVLKRVMKEHLHRAYKI-----KEWENE------NDFLLQLCKLTGKL 352
+ + + + V RV + + Y I K +E++ ++ L C G+
Sbjct: 183 IDRMNQHRECVQVVANRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRGQT 362
Query: 353 LKGGEPDLMTAAKMVLHDWQRGKIPFFVPPP-------QQEDLSEEPIVNGLDVDDGVDS 405
G PD A++ +L D+ GK+P + PP ED +E VN V D D
Sbjct: 363 TSSGLPDETRASRQILKDYIDGKLPHYEMPPGLSTQELASEDSNEHDQVNP-HVSDASDI 539
Query: 406 NQASAAIKAIA 416
+S +A
Sbjct: 540 EDSSVVETELA 572
>TC90237 similar to GP|20466544|gb|AAM20589.1 unknown protein {Arabidopsis
thaliana}, partial (79%)
Length = 1174
Score = 45.1 bits (105), Expect = 6e-05
Identities = 56/207 (27%), Positives = 88/207 (42%), Gaps = 21/207 (10%)
Frame = +1
Query: 137 SDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMMLLLNKCDLIPAWATKGWLRVL-SKE 195
+D+V++V DAR P + L+ HL K ++ LNK DL W+ S +
Sbjct: 61 ADLVIEVRDARIPISSINSDLQPHLS----LKRRVVALNKKDLANPNIMHKWVNYFESCK 228
Query: 196 YPTLAFHASINKSFGKGSLLSVLRQFARLKSDKQAIS------VGFIGYPNVGKSSVINT 249
+ +A S K L + LK K+AIS V +G PNVGKS +IN+
Sbjct: 229 QDCIPINAHSKSSVTK------LLELVELKL-KEAISKEPTLLVMVVGVPNVGKSCLINS 387
Query: 250 L-----------RTKNVCKVAPIPGETKVWQYITLTKR--IFLIDCPGVVYQN-KDSEAD 295
+ V P+PG T+ + + I+++D PGV+ + D E
Sbjct: 388 IHQIAHSRFPVQEKMKRAAVGPLPGVTQDIAGFKIANKPSIYVLDTPGVLVPSISDIETG 567
Query: 296 VVLKGVVRVTNLKDAADRIGEVLKRVM 322
+ L V + +RI + L V+
Sbjct: 568 LKLALAGSVKDSVVGEERIAQYLLAVL 648
>TC81248 similar to SP|O81004|YB87_ARATH Hypothetical GTP-binding protein
At2g22870. [Mouse-ear cress] {Arabidopsis thaliana},
partial (78%)
Length = 1177
Score = 43.5 bits (101), Expect = 2e-04
Identities = 28/84 (33%), Positives = 45/84 (53%), Gaps = 3/84 (3%)
Frame = +3
Query: 216 SVLRQFARLKSDKQAISVG--FIGYPNVGKSSVINTL-RTKNVCKVAPIPGETKVWQYIT 272
S LR RL+ + I + F+ NVGKSS+IN+L R K + + PG+T++ +
Sbjct: 345 SSLRVVRRLRIAPKIIDLNSLFLVVSNVGKSSLINSLVRKKELALTSKKPGKTQLINHFL 524
Query: 273 LTKRIFLIDCPGVVYQNKDSEADV 296
+ K+ +L+D PG + A V
Sbjct: 525 VNKKWYLVDLPGYGFAKASEAAKV 596
>TC88009 similar to PIR|T48507|T48507 probable GTP-binding protein -
Arabidopsis thaliana, partial (72%)
Length = 1399
Score = 38.9 bits (89), Expect = 0.004
Identities = 19/59 (32%), Positives = 34/59 (57%), Gaps = 1/59 (1%)
Frame = +1
Query: 236 IGYPNVGKSSVINTL-RTKNVCKVAPIPGETKVWQYITLTKRIFLIDCPGVVYQNKDSE 293
+G NVGKSS++N++ R K + + PG+T++ + + +L+D PG Y + E
Sbjct: 640 VGRSNVGKSSLLNSIVRRKKLALTSKKPGKTQLINHFRVNDSWYLVDLPGYGYASAPQE 816
>BI272296 similar to GP|12322014|gb GTPase putative; 34281-30152
{Arabidopsis thaliana}, partial (22%)
Length = 487
Score = 35.0 bits (79), Expect = 0.064
Identities = 15/35 (42%), Positives = 24/35 (67%)
Frame = +2
Query: 232 SVGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETK 266
++ +G PNVGKSS++N L ++ V+PI G T+
Sbjct: 179 AISIVGRPNVGKSSILNALVGEDRTIVSPISGTTR 283
>TC88185 homologue to PIR|T09368|T09368 GTP-binding protein F23K16.150 -
Arabidopsis thaliana, complete
Length = 1734
Score = 34.7 bits (78), Expect = 0.083
Identities = 21/71 (29%), Positives = 36/71 (50%), Gaps = 10/71 (14%)
Frame = +3
Query: 233 VGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLT----------KRIFLIDC 282
VG +G+P+VGKS+++N K+ E +++ TLT +I L+D
Sbjct: 348 VGLVGFPSVGKSTLLN--------KLTGTFSEVASYEFTTLTCIPGVITYRGAKIQLLDL 503
Query: 283 PGVVYQNKDSE 293
PG++ KD +
Sbjct: 504 PGIIEGAKDGK 536
>TC87351 similar to GP|10176988|dbj|BAB10220. GTP-binding protein-like
{Arabidopsis thaliana}, partial (81%)
Length = 2266
Score = 34.3 bits (77), Expect = 0.11
Identities = 39/159 (24%), Positives = 66/159 (40%), Gaps = 25/159 (15%)
Frame = +3
Query: 133 VIDSSDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMMLLLNKCDLIPAWATKGWLRVL 192
V+ +S ++ + DAR + K L++N +L++NK + + + G L
Sbjct: 597 VLSTSHSLLFLTDARAGLHPLDQEVGKWLRKNAPQLKPILVMNKSESL--FDVDGSLASS 770
Query: 193 SKEYPTLAFHASINKSFGKG--------SLLSVLRQFARLKSDKQ--------------- 229
+ E L F I S G SL VL + +S K+
Sbjct: 771 ANEMSRLGFGDPIAISAETGLGMHDLFLSLQPVLEDYMLNESAKENTCGDESSFPEADES 950
Query: 230 --AISVGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETK 266
+ + +G PNVGKS+++NTL ++ V P G T+
Sbjct: 951 KLPLQLAIVGRPNVGKSTLLNTLLQEDRVLVGPEAGLTR 1067
>TC86616 similar to GP|7643796|gb|AAF65513.1| GTP-binding protein {Capsicum
annuum}, partial (57%)
Length = 886
Score = 30.8 bits (68), Expect = 1.2
Identities = 11/20 (55%), Positives = 16/20 (80%)
Frame = +2
Query: 231 ISVGFIGYPNVGKSSVINTL 250
+ +G +G PNVGKS++ NTL
Sbjct: 179 LKIGIVGLPNVGKSTLFNTL 238
>TC82574 similar to GP|17473914|gb|AAL38371.1 GTP-binding protein-like
{Arabidopsis thaliana}, partial (54%)
Length = 915
Score = 30.8 bits (68), Expect = 1.2
Identities = 26/106 (24%), Positives = 47/106 (43%), Gaps = 13/106 (12%)
Frame = +2
Query: 233 VGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLT---KRIFLIDCPGVVYQN 289
V +G PNVGKS++ N + + + V P T+ + ++ L D PGV+ +
Sbjct: 350 VALLGKPNVGKSTLANQMVGQKLSIVTDKPQTTRHRVLCICSGSDYQMVLYDTPGVLQEQ 529
Query: 290 KDSEADVVLKGVVRVTNLKD----------AADRIGEVLKRVMKEH 325
+ ++++ V T D A ++I EVL+ + H
Sbjct: 530 RHKLDSMMMQNVRSATVNADCVLVLVDACKAPEKIDEVLEEAIGGH 667
>AW688430 similar to GP|22535581|dbj contains EST AU182189(C63973)~similar to
Arabidopsis thaliana chromosome5 At5g65910~unknown,
partial (2%)
Length = 674
Score = 30.0 bits (66), Expect = 2.1
Identities = 13/40 (32%), Positives = 23/40 (57%)
Frame = +2
Query: 385 QEDLSEEPIVNGLDVDDGVDSNQASAAIKAIANVLSSQQQ 424
QE+ EE +VN + +DD D + A+ + + ++QQQ
Sbjct: 68 QEEEEEELVVNKVQIDDDDDDDAAAVTVTGNNKIAAAQQQ 187
>TC85558 homologue to GP|7453538|gb|AAF62870.1| Toc64 {Pisum sativum},
partial (40%)
Length = 1387
Score = 30.0 bits (66), Expect = 2.1
Identities = 20/75 (26%), Positives = 35/75 (46%), Gaps = 2/75 (2%)
Frame = +3
Query: 278 FLIDCPGVVYQNKDSEADVVL--KGVVRVTNLKDAADRIGEVLKRVMKEHLHRAYKIKEW 335
FL+D +Y + +AD+ K V + + +A+ + KE ++AYK K+W
Sbjct: 471 FLLDTLKTMYTSLQEQADIAATSKASRNVVSKEQSAE--------IAKEKGNQAYKDKQW 626
Query: 336 ENENDFLLQLCKLTG 350
+ F + KL G
Sbjct: 627 QKAIGFYTEAIKLCG 671
>TC91051 similar to PIR|H84787|H84787 probable receptor-like protein kinase
[imported] - Arabidopsis thaliana, partial (36%)
Length = 1240
Score = 29.6 bits (65), Expect = 2.7
Identities = 12/34 (35%), Positives = 23/34 (67%)
Frame = +3
Query: 387 DLSEEPIVNGLDVDDGVDSNQASAAIKAIANVLS 420
D S+ P+VN ++++ ++ N S ++AI+ VLS
Sbjct: 1074 DSSKGPLVNAMEINKYLEKNDGSPDVEAISGVLS 1175
>TC78598 similar to SP|Q43207|FKB7_WHEAT 70 kDa peptidylprolyl isomerase (EC
5.2.1.8) (Peptidylprolyl cis-trans isomerase) (PPiase),
partial (92%)
Length = 2028
Score = 28.9 bits (63), Expect = 4.6
Identities = 38/132 (28%), Positives = 56/132 (41%), Gaps = 5/132 (3%)
Frame = +2
Query: 313 RIGEVLKRVMKEHLHRAYKIKEWENENDFLLQLCKLTGKLLKGGEPDLMTAAKMVLHDWQ 372
RIG L+ +KE + +E ND + KLTGKL G T HD
Sbjct: 992 RIGRALRETLKEG-------EGYERPNDGAVVQVKLTGKLQDG------TVFFKKGHD-- 1126
Query: 373 RGKIPFFVPPPQQEDLSEEPIVNGLD--VDDGVDSNQASAAIK---AIANVLSSQQQSSV 427
G PF + + EE +++GLD V + A I+ A + S Q+ ++V
Sbjct: 1127 -GDQPF------EFRIDEEQVIDGLDRAVKNMKKGEIALVIIQPEYAFGSSGSPQELTTV 1285
Query: 428 PVQKDLFTENEL 439
P ++ E EL
Sbjct: 1286 PPNSTVYYEVEL 1321
>CB891834 homologue to PIR|C96751|C96 probable GTP-binding protein F28P22.15
[imported] - Arabidopsis thaliana, partial (43%)
Length = 518
Score = 28.5 bits (62), Expect = 6.0
Identities = 18/64 (28%), Positives = 31/64 (48%), Gaps = 10/64 (15%)
Frame = +3
Query: 233 VGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLT----------KRIFLIDC 282
V IG+P+VGKS+++ L + E +++ TLT +I L+D
Sbjct: 174 VSLIGFPSVGKSTLLTLLTGTH--------SEAASYEFTTLTCIPGIIHYNDTKIQLLDL 329
Query: 283 PGVV 286
PG++
Sbjct: 330 PGII 341
>BF640447 similar to GP|20152085|gb| RE21802p {Drosophila melanogaster},
partial (15%)
Length = 326
Score = 28.5 bits (62), Expect = 6.0
Identities = 11/20 (55%), Positives = 14/20 (70%)
Frame = +2
Query: 231 ISVGFIGYPNVGKSSVINTL 250
+ VG +G PNVGKS+ N L
Sbjct: 209 LKVGIVGVPNVGKSTFFNVL 268
>TC88979 similar to PIR|H96601|H96601 hypothetical protein T6H22.14
[imported] - Arabidopsis thaliana, partial (89%)
Length = 1616
Score = 28.5 bits (62), Expect = 6.0
Identities = 11/33 (33%), Positives = 18/33 (54%)
Frame = +3
Query: 230 AISVGFIGYPNVGKSSVINTLRTKNVCKVAPIP 262
++ G +G PNVGKS++ N + + A P
Sbjct: 201 SLKAGIVGLPNVGKSTLFNAVVENGKAQAANFP 299
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.132 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,397,892
Number of Sequences: 36976
Number of extensions: 175497
Number of successful extensions: 730
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 717
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 725
length of query: 482
length of database: 9,014,727
effective HSP length: 100
effective length of query: 382
effective length of database: 5,317,127
effective search space: 2031142514
effective search space used: 2031142514
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0030.10


