

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0029b.5
(313 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
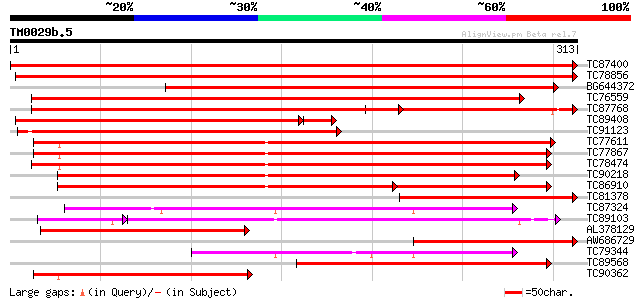
Score E
Sequences producing significant alignments: (bits) Value
TC87400 SP|Q06009|P2A_MEDSA Serine/threonine protein phosphatase... 639 0.0
TC78856 GP|7248359|dbj|BAA92697.1 type 2A protein phosphatase-1 ... 529 e-151
BG644372 homologue to SP|O04860|P2A5 Serine/threonine protein ph... 440 e-124
TC76559 GP|20385063|gb|AAM21172.1 serine/threonine protein phosp... 365 e-102
TC87768 homologue to SP|P48528|PPX2_ARATH Serine/threonine prote... 317 5e-87
TC89408 homologue to SP|Q9XGH7|P2A_TOBAC Serine/threonine protei... 297 2e-85
TC91123 homologue to GP|7248361|dbj|BAA92698.1 type 2A protein p... 302 1e-82
TC77611 SP|P48488|PP1_MEDVA Serine/threonine protein phosphatase... 291 2e-79
TC77867 PIR|T09544|T09544 phosphoprotein phosphatase (EC 3.1.3.1... 289 1e-78
TC78474 homologue to PIR|T09550|T09550 phosphoprotein phosphatas... 280 4e-76
TC90218 homologue to SP|P48490|PP1_PHAVU Serine/threonine protei... 273 6e-74
TC86910 homologue to PIR|T09548|T09548 phosphoprotein phosphatas... 217 2e-71
TC81378 homologue to GP|19071628|gb|AAL84295.1 serine/threonine ... 206 9e-54
TC87324 homologue to PIR|E86217|E86217 protein T27G7.10 [importe... 198 3e-51
TC89103 similar to GP|16930441|gb|AAL31906.1 At2g42810/F7D19.19 ... 162 3e-42
AL378129 GP|20385063|g serine/threonine protein phosphatase 2A {... 160 5e-40
AW686729 GP|7248361|dbj type 2A protein phosphatase-2 {Vicia fab... 147 7e-36
TC79344 homologue to PIR|E86217|E86217 protein T27G7.10 [importe... 139 2e-33
TC89568 homologue to PIR|T09544|T09544 phosphoprotein phosphatas... 139 2e-33
TC90362 homologue to PIR|T09547|T09547 phosphoprotein phosphatas... 124 4e-29
>TC87400 SP|Q06009|P2A_MEDSA Serine/threonine protein phosphatase PP2A
catalytic subunit (EC 3.1.3.16). [Alfalfa], complete
Length = 1439
Score = 639 bits (1649), Expect = 0.0
Identities = 303/313 (96%), Positives = 309/313 (97%)
Frame = +1
Query: 1 MGANSLPSESTHDLDEQISQLMQCKPLSEQQVKELCEKAKEILMEESNVQPVKSPVTICG 60
MGANS+ ++ THDL+EQISQLMQCKPLSEQQVKELCEKAKEILM+ESNVQPVKSPVTICG
Sbjct: 91 MGANSMIADVTHDLNEQISQLMQCKPLSEQQVKELCEKAKEILMDESNVQPVKSPVTICG 270
Query: 61 DIHGQFHDLAELFRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLVALKVRYHQRITILRG 120
DIHGQFHDLAELFRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLVALKVRY QRITILRG
Sbjct: 271 DIHGQFHDLAELFRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLVALKVRYPQRITILRG 450
Query: 121 NHESRQITQVYGFYDECLRKYGNANVWKTFTDLFDFFPLTALVESEIFCLHGGLSPSIET 180
NHESRQITQVYGFYDECLRKYG+ANVWK FTDLFDFFPLTALVESEIFCLHGGLSPSIET
Sbjct: 451 NHESRQITQVYGFYDECLRKYGSANVWKIFTDLFDFFPLTALVESEIFCLHGGLSPSIET 630
Query: 181 LDNIRNFDRVQEVPHEGPMCDLLWSDPDDRCGWGISPRGAGYTFGQDISEQFNHTNSLKL 240
LDNIRNFDRVQEVPHEGPMCDLLWSDPDDRCGWGISPRGAGYTFGQDISEQFNHTNSLKL
Sbjct: 631 LDNIRNFDRVQEVPHEGPMCDLLWSDPDDRCGWGISPRGAGYTFGQDISEQFNHTNSLKL 810
Query: 241 IARAHQLVMDGFNWAHEQKVVTIFSAPNYCYRCGNMASILEVDDCKGHTFIQFEPAPRRG 300
IARAHQLVMDGFNWAHEQKVVTIFSAPNYCYRCGNMASILEVDDCKGHTFIQFEPAPRRG
Sbjct: 811 IARAHQLVMDGFNWAHEQKVVTIFSAPNYCYRCGNMASILEVDDCKGHTFIQFEPAPRRG 990
Query: 301 EPDVTRRTPDYFL 313
EPDVTRRTPDYFL
Sbjct: 991 EPDVTRRTPDYFL 1029
>TC78856 GP|7248359|dbj|BAA92697.1 type 2A protein phosphatase-1 {Vicia
faba}, complete
Length = 1426
Score = 529 bits (1362), Expect = e-151
Identities = 244/311 (78%), Positives = 274/311 (87%), Gaps = 1/311 (0%)
Frame = +3
Query: 4 NSLPSESTH-DLDEQISQLMQCKPLSEQQVKELCEKAKEILMEESNVQPVKSPVTICGDI 62
N+L + +H DLD QI LM+CKPL E VK LC++A+ IL+EE NVQPVK PVT+CGDI
Sbjct: 174 NNLSTMPSHADLDRQIESLMECKPLPEADVKALCDQARAILVEEWNVQPVKCPVTVCGDI 353
Query: 63 HGQFHDLAELFRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNH 122
HGQF+DL ELFRIGG PDTNYLFMGDYVDRGYYSVETV+LLVALKVRY RITILRGNH
Sbjct: 354 HGQFYDLIELFRIGGNAPDTNYLFMGDYVDRGYYSVETVSLLVALKVRYRDRITILRGNH 533
Query: 123 ESRQITQVYGFYDECLRKYGNANVWKTFTDLFDFFPLTALVESEIFCLHGGLSPSIETLD 182
ESRQITQVYGFYDECLRKYGNANVWK FTDLFD+ PLTAL+ES+IFCLHGGLSPS++TLD
Sbjct: 534 ESRQITQVYGFYDECLRKYGNANVWKFFTDLFDYLPLTALIESQIFCLHGGLSPSLDTLD 713
Query: 183 NIRNFDRVQEVPHEGPMCDLLWSDPDDRCGWGISPRGAGYTFGQDISEQFNHTNSLKLIA 242
NIR DR+QEVPHEGPMCDLLWSDPDDRCGWGISPRGAGYTFGQDI+ QFNHTN L LI+
Sbjct: 714 NIRALDRIQEVPHEGPMCDLLWSDPDDRCGWGISPRGAGYTFGQDIAAQFNHTNGLSLIS 893
Query: 243 RAHQLVMDGFNWAHEQKVVTIFSAPNYCYRCGNMASILEVDDCKGHTFIQFEPAPRRGEP 302
RAHQLVM+G+NW E+ VVT+FSAPNYCYRCGNMA+ILE+ + F+QF+PAPR+ EP
Sbjct: 894 RAHQLVMEGYNWCQEKNVVTVFSAPNYCYRCGNMAAILEIGENMDQNFLQFDPAPRQIEP 1073
Query: 303 DVTRRTPDYFL 313
D TR+TPDYFL
Sbjct: 1074DTTRKTPDYFL 1106
>BG644372 homologue to SP|O04860|P2A5 Serine/threonine protein phosphatase
PP2A-5 catalytic subunit (EC 3.1.3.16). [Common
tobacco], partial (69%)
Length = 658
Score = 440 bits (1131), Expect = e-124
Identities = 202/217 (93%), Positives = 214/217 (98%)
Frame = +1
Query: 87 MGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHESRQITQVYGFYDECLRKYGNANV 146
MGDYVDRGYYSVETVTLLVALKVRY QR+TILRGNHESRQITQVYGFYDECLRKYGNANV
Sbjct: 4 MGDYVDRGYYSVETVTLLVALKVRYPQRLTILRGNHESRQITQVYGFYDECLRKYGNANV 183
Query: 147 WKTFTDLFDFFPLTALVESEIFCLHGGLSPSIETLDNIRNFDRVQEVPHEGPMCDLLWSD 206
WKTFTDLFD+FPLTALVESEIFCLHGGLSPSIETLDN+R+FDRVQEVPHEG MCDLLWSD
Sbjct: 184 WKTFTDLFDYFPLTALVESEIFCLHGGLSPSIETLDNVRSFDRVQEVPHEGAMCDLLWSD 363
Query: 207 PDDRCGWGISPRGAGYTFGQDISEQFNHTNSLKLIARAHQLVMDGFNWAHEQKVVTIFSA 266
PDDRCGWG+SPRGAGYTFGQDISEQF+HTN+LKLIARAHQLVM+G+NW+HEQKVVTIFSA
Sbjct: 364 PDDRCGWGMSPRGAGYTFGQDISEQFHHTNNLKLIARAHQLVMEGYNWSHEQKVVTIFSA 543
Query: 267 PNYCYRCGNMASILEVDDCKGHTFIQFEPAPRRGEPD 303
PNYCYRCGNMASILE DDC+GHTFIQF+PAPRRGEPD
Sbjct: 544 PNYCYRCGNMASILESDDCRGHTFIQFDPAPRRGEPD 654
>TC76559 GP|20385063|gb|AAM21172.1 serine/threonine protein phosphatase 2A
{Pisum sativum}, complete
Length = 1305
Score = 365 bits (938), Expect = e-102
Identities = 164/273 (60%), Positives = 213/273 (77%), Gaps = 1/273 (0%)
Frame = +2
Query: 13 DLDEQISQLMQCKPLSEQQVKELCEKAKEILMEESNVQPVKSPVTICGDIHGQFHDLAEL 72
DLD+ IS++ + L E +++ LCE KEIL+EESNVQPV SPVT+CGDIHGQFHDL +L
Sbjct: 134 DLDQWISKVKDGQHLLEDELQLLCEYVKEILIEESNVQPVNSPVTVCGDIHGQFHDLMKL 313
Query: 73 FRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHESRQITQVYG 132
F+ GG P+TNY+FMGD+VDRGY S+E T+L+ LK RY IT+LRGNHESRQ+TQVYG
Sbjct: 314 FQTGGHVPETNYIFMGDFVDRGYNSLEVFTILLLLKARYPANITLLRGNHESRQLTQVYG 493
Query: 133 FYDECLRKYGNANVWKTFTDLFDFFPLTALVESEIFCLHGGLSPSIETLDNIRNFDRVQE 192
FYDEC RKYGNAN W+ TD+FD+ L+A+++ + C+HGGLSP I T+D IR +R E
Sbjct: 494 FYDECQRKYGNANAWRYCTDVFDYLTLSAIIDGTVLCVHGGLSPDIRTIDQIRVIERNCE 673
Query: 193 VPHEGPMCDLLWSDPDDRCGWGISPRGAGYTFGQDISEQFNHTNSLKLIARAHQLVMDGF 252
+PHEGP CDL+WSDP+D W +SPRGAG+ FG ++ +FNH N+L L+ RAHQLV +G
Sbjct: 674 IPHEGPFCDLMWSDPEDIETWAVSPRGAGWLFGSRVTSEFNHINNLDLVCRAHQLVQEGL 853
Query: 253 NWAHEQK-VVTIFSAPNYCYRCGNMASILEVDD 284
+ + K +VT++SAPNYCYRCGN+ASIL ++
Sbjct: 854 KYMFQDKGLVTVWSAPNYCYRCGNVASILSFNE 952
>TC87768 homologue to SP|P48528|PPX2_ARATH Serine/threonine protein
phosphatase PP-X isozyme 2 (EC 3.1.3.16). [Mouse-ear
cress], complete
Length = 1430
Score = 317 bits (811), Expect = 5e-87
Identities = 146/206 (70%), Positives = 174/206 (83%), Gaps = 1/206 (0%)
Frame = +3
Query: 13 DLDEQISQLMQCKPLSEQQVKELCEKAKEILMEESNVQPVKSPVTICGDIHGQFHDLAEL 72
DLD QI QL +C+PL E +VK LC KA EIL+EESNVQ V +PVTICGDIHGQF+D+ EL
Sbjct: 144 DLDRQIEQLKRCEPLKESEVKSLCLKAIEILVEESNVQRVDAPVTICGDIHGQFYDMKEL 323
Query: 73 FRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHESRQITQVYG 132
F++GG CP TNYLF+GD+VDRG+YSVET LL+ALKVRY RIT++RGNHESRQITQVYG
Sbjct: 324 FKVGGDCPKTNYLFLGDFVDRGFYSVETFLLLLALKVRYPDRITLIRGNHESRQITQVYG 503
Query: 133 FYDECLRKYGNANVWKTFTDLFDFFPLTALVESEIFCLHGGLSPSIETLDNIRNFDRVQE 192
FYDECLRKYG+ NVW+ TD+FD+ L+AL+E++IF +HGGLSP+I TLD IR DR QE
Sbjct: 504 FYDECLRKYGSVNVWRYCTDIFDYLSLSALIENKIFSVHGGLSPAISTLDQIRTIDRKQE 683
Query: 193 VPHEGPMCDLLWSDPDDRC-GWGISP 217
VPH+G MCDLLWSDP+D GWG+SP
Sbjct: 684 VPHDGAMCDLLWSDPEDIVDGWGLSP 761
Score = 111 bits (278), Expect = 3e-25
Identities = 57/120 (47%), Positives = 76/120 (62%), Gaps = 3/120 (2%)
Frame = +1
Query: 197 GPMCDLLWSDPDDRCGWGI-SPRGAGYTFGQDISEQFNHTNSLKLIARAHQLVMDGFNWA 255
G + +LW D G+ PRGAG+ FG + FNHTN++ I RAHQLVM+G+ W
Sbjct: 718 GQILKILWMD-------GV*VPRGAGFLFGGSVVTGFNHTNNIDYICRAHQLVMEGYKWM 876
Query: 256 HEQKVVTIFSAPNYCYRCGNMASILEVDDCKGHTFIQFEPAPR--RGEPDVTRRTPDYFL 313
++VT++SAPNYCYRCGN+A+ILE+D F F+ AP+ RG P + PDYFL
Sbjct: 877 FNNQIVTVWSAPNYCYRCGNVAAILELDGNLNKQFRVFDAAPQESRGAP-AKKPAPDYFL 1053
>TC89408 homologue to SP|Q9XGH7|P2A_TOBAC Serine/threonine protein
phosphatase PP2A catalytic subunit (EC 3.1.3.16).
[Common tobacco], partial (55%)
Length = 656
Score = 297 bits (761), Expect(2) = 2e-85
Identities = 142/159 (89%), Positives = 152/159 (95%)
Frame = +1
Query: 4 NSLPSESTHDLDEQISQLMQCKPLSEQQVKELCEKAKEILMEESNVQPVKSPVTICGDIH 63
+++ S S +LDEQISQLMQCKPLSEQ+V+ LC+KAKEILMEESNVQPVKSPVTICGDIH
Sbjct: 124 SNMDSNSHGNLDEQISQLMQCKPLSEQEVRVLCDKAKEILMEESNVQPVKSPVTICGDIH 303
Query: 64 GQFHDLAELFRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHE 123
GQFHDLAELFRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLV+LKVRY QRITILRGNHE
Sbjct: 304 GQFHDLAELFRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLVSLKVRYRQRITILRGNHE 483
Query: 124 SRQITQVYGFYDECLRKYGNANVWKTFTDLFDFFPLTAL 162
SRQITQVYGFYDECLRKYG+ANVWK FTDLFD+FPLTAL
Sbjct: 484 SRQITQVYGFYDECLRKYGSANVWKMFTDLFDYFPLTAL 600
Score = 36.6 bits (83), Expect(2) = 2e-85
Identities = 15/18 (83%), Positives = 17/18 (94%)
Frame = +2
Query: 163 VESEIFCLHGGLSPSIET 180
VESEIFCLHGGL+P +ET
Sbjct: 602 VESEIFCLHGGLTPYLET 655
>TC91123 homologue to GP|7248361|dbj|BAA92698.1 type 2A protein
phosphatase-2 {Vicia faba}, partial (57%)
Length = 690
Score = 302 bits (774), Expect = 1e-82
Identities = 142/179 (79%), Positives = 161/179 (89%)
Frame = +2
Query: 5 SLPSESTHDLDEQISQLMQCKPLSEQQVKELCEKAKEILMEESNVQPVKSPVTICGDIHG 64
S+PS++ DLD QI LM+CKPL+E +VK LC++A+ IL+EE NVQPVK PVT+CGDIHG
Sbjct: 158 SMPSQA--DLDRQIEHLMECKPLTEAEVKALCDQARAILVEEWNVQPVKCPVTVCGDIHG 331
Query: 65 QFHDLAELFRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHES 124
QF+DL ELFRIGG P TNYLFMGDYVDRGYYSVETV+LLVALKVRY RITILRGNHES
Sbjct: 332 QFYDLIELFRIGGNAPHTNYLFMGDYVDRGYYSVETVSLLVALKVRYRDRITILRGNHES 511
Query: 125 RQITQVYGFYDECLRKYGNANVWKTFTDLFDFFPLTALVESEIFCLHGGLSPSIETLDN 183
RQITQVYGFYDECLRKYGNANVWK FTDLFD+ PLTAL+ES++FCLHGGLSPS++TLDN
Sbjct: 512 RQITQVYGFYDECLRKYGNANVWKFFTDLFDYLPLTALIESQVFCLHGGLSPSLDTLDN 688
>TC77611 SP|P48488|PP1_MEDVA Serine/threonine protein phosphatase PP1 (EC
3.1.3.16). {Medicago varia}, complete
Length = 1509
Score = 291 bits (745), Expect = 2e-79
Identities = 139/297 (46%), Positives = 197/297 (65%), Gaps = 9/297 (3%)
Frame = +2
Query: 14 LDEQISQLMQCKP--------LSEQQVKELCEKAKEILMEESNVQPVKSPVTICGDIHGQ 65
LD+ I +L+ K L+E +++LC AKEI + + N+ +++P+ ICGD+HGQ
Sbjct: 200 LDDIIKKLVSAKNGRTTKQVHLTEADIRQLCTSAKEIFLSQPNLLELEAPIKICGDVHGQ 379
Query: 66 FHDLAELFRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHESR 125
F DL LF GG P+ NYLF+GDYVDRG S+ET+ LL+A K++Y + +LRGNHE
Sbjct: 380 FSDLLRLFEYGGYPPEANYLFLGDYVDRGKQSIETICLLLAYKIKYKENFFLLRGNHECA 559
Query: 126 QITQVYGFYDECLRKYGNANVWKTFTDLFDFFPLTALVESEIFCLHGGLSPSIETLDNIR 185
I ++YGFYDEC R+Y N +WKTFTD F+ P+ ALV+ +I C+HGGLSP ++ LD IR
Sbjct: 560 SINRIYGFYDECKRRY-NVRLWKTFTDCFNCLPVAALVDEKILCMHGGLSPELKNLDQIR 736
Query: 186 NFDRVQEVPHEGPMCDLLWSDPD-DRCGWGISPRGAGYTFGQDISEQFNHTNSLKLIARA 244
N R +VP G +CDLLW+DPD D GWG + RG +TFG D +F + L LI RA
Sbjct: 737 NIARPIDVPDHGLLCDLLWADPDKDLEGWGENDRGVSFTFGADKVVEFLEHHDLDLICRA 916
Query: 245 HQLVMDGFNWAHEQKVVTIFSAPNYCYRCGNMASILEVDDCKGHTFIQFEPAPRRGE 301
HQ+V DG+ + ++K+VT+FSAPNYC N +++ VDD +F + + ++G+
Sbjct: 917 HQVVEDGYEFFAKRKLVTVFSAPNYCGEFDNAGAMMSVDDSLTCSFQILKSSDKKGK 1087
>TC77867 PIR|T09544|T09544 phosphoprotein phosphatase (EC 3.1.3.16)
catalytic beta chain - alfalfa, complete
Length = 1583
Score = 289 bits (739), Expect = 1e-78
Identities = 135/294 (45%), Positives = 195/294 (65%), Gaps = 8/294 (2%)
Frame = +3
Query: 14 LDEQISQLMQCKP-------LSEQQVKELCEKAKEILMEESNVQPVKSPVTICGDIHGQF 66
LD+ I++L++ + LSE ++++LC A+EI +++ N+ +++P+ ICGDIHGQ+
Sbjct: 246 LDDIINRLLEVRSRPGKQVQLSETEIRQLCSTAREIFLQQPNLLELEAPIKICGDIHGQY 425
Query: 67 HDLAELFRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHESRQ 126
DL LF GG P NYLF+GDYVDRG S+ET+ LL+A K++Y + +LRGNHE
Sbjct: 426 SDLLRLFEYGGLPPQANYLFLGDYVDRGKQSLETICLLLAYKIKYPENFFLLRGNHECAS 605
Query: 127 ITQVYGFYDECLRKYGNANVWKTFTDLFDFFPLTALVESEIFCLHGGLSPSIETLDNIRN 186
I ++YGFYDEC R++ N +WK FTD F+ P+ AL++ +I C+HGGLSP + LD IRN
Sbjct: 606 INRIYGFYDECKRRF-NVRIWKVFTDCFNCLPVAALIDEKILCMHGGLSPDLHNLDQIRN 782
Query: 187 FDRVQEVPHEGPMCDLLWSDPDDRC-GWGISPRGAGYTFGQDISEQFNHTNSLKLIARAH 245
R +VP G +CDLLWSDP GWG++ RG YTFG D +F + L LI RAH
Sbjct: 783 LQRPTDVPDTGLLCDLLWSDPSKEVQGWGMNDRGVSYTFGADKVSEFLQKHDLDLICRAH 962
Query: 246 QLVMDGFNWAHEQKVVTIFSAPNYCYRCGNMASILEVDDCKGHTFIQFEPAPRR 299
Q+V DG+ + +++VTIFSAPNYC N +++ VD+ +F +PA ++
Sbjct: 963 QVVEDGYEFFANRQLVTIFSAPNYCGEFDNAGAMMSVDETLMCSFQILKPADKK 1124
>TC78474 homologue to PIR|T09550|T09550 phosphoprotein phosphatase (EC
3.1.3.16) 1 catalytic epsilon chain - alfalfa, complete
Length = 1597
Score = 280 bits (717), Expect = 4e-76
Identities = 131/293 (44%), Positives = 196/293 (66%), Gaps = 6/293 (2%)
Frame = +3
Query: 13 DLDEQISQLMQCKP-----LSEQQVKELCEKAKEILMEESNVQPVKSPVTICGDIHGQFH 67
D+ +++++ +P LSE ++K+LC +++I +++ N+ +++P+ ICGDIHGQ+
Sbjct: 201 DIIRRLTEVRLSRPGKQVQLSEAEIKQLCSASRDIFLQQPNLLELEAPIKICGDIHGQYS 380
Query: 68 DLAELFRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHESRQI 127
DL LF GG P +NYLF+GDYVDRG S+ET+ LL+A K++Y + +LRGNHE I
Sbjct: 381 DLLRLFEYGGLPPQSNYLFLGDYVDRGKQSLETICLLLAYKIKYPENFFLLRGNHECASI 560
Query: 128 TQVYGFYDECLRKYGNANVWKTFTDLFDFFPLTALVESEIFCLHGGLSPSIETLDNIRNF 187
++YGFYDEC R++ N +WK FT+ F+ P+ AL++ +I C+HGGLSP + LD IRN
Sbjct: 561 NRIYGFYDECKRRF-NVRLWKAFTESFNCLPVAALIDEKILCMHGGLSPDLTNLDQIRNL 737
Query: 188 DRVQEVPHEGPMCDLLWSDP-DDRCGWGISPRGAGYTFGQDISEQFNHTNSLKLIARAHQ 246
R +P G +CDLLWSDP D GWG++ RG YTFG D +F + L LI RAHQ
Sbjct: 738 PRPVPIPDTGLLCDLLWSDPGKDVKGWGMNDRGVSYTFGPDKVAEFLTRHDLDLICRAHQ 917
Query: 247 LVMDGFNWAHEQKVVTIFSAPNYCYRCGNMASILEVDDCKGHTFIQFEPAPRR 299
+V DG+ + ++++VTIFSAPNYC N +++ VD+ +F +PA ++
Sbjct: 918 VVEDGYEFFADRQLVTIFSAPNYCGEFDNAGAMMSVDENLMCSFQILKPAEKK 1076
>TC90218 homologue to SP|P48490|PP1_PHAVU Serine/threonine protein
phosphatase PP1 (EC 3.1.3.16). [Kidney bean French
bean], partial (95%)
Length = 1024
Score = 273 bits (698), Expect = 6e-74
Identities = 125/256 (48%), Positives = 174/256 (67%), Gaps = 1/256 (0%)
Frame = +2
Query: 27 LSEQQVKELCEKAKEILMEESNVQPVKSPVTICGDIHGQFHDLAELFRIGGKCPDTNYLF 86
LSE ++++LC A++I + + + +++P+ ICGDIHGQ+ DL LF GG P NYLF
Sbjct: 179 LSESEIRQLCINARQIFLSQPILLDLRAPIRICGDIHGQYQDLLRLFEYGGYPPAANYLF 358
Query: 87 MGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHESRQITQVYGFYDECLRKYGNANV 146
+GDYVDRG S+ET+ LL+A K+RY R+ +LRGNHE +I ++YGFYDEC R++ N +
Sbjct: 359 LGDYVDRGKQSLETICLLLAYKIRYPDRVYLLRGNHEDAKINRIYGFYDECKRRF-NVRL 535
Query: 147 WKTFTDLFDFFPLTALVESEIFCLHGGLSPSIETLDNIRNFDRVQEVPHEGPMCDLLWSD 206
WK FTD F+ P+ AL++ +I C+HGGLSP ++ LD IR R E+P G +CDLLWSD
Sbjct: 536 WKIFTDCFNCLPVAALIDEKILCMHGGLSPELQNLDQIREVTRPTEIPDSGLLCDLLWSD 715
Query: 207 PDDRC-GWGISPRGAGYTFGQDISEQFNHTNSLKLIARAHQLVMDGFNWAHEQKVVTIFS 265
PD + GW S RG TFG D+ +F N L LI R HQ+V DG+ + ++++VTIFS
Sbjct: 716 PDPKAEGWADSDRGVSCTFGSDVVAEFLDKNDLDLICRGHQVVEDGYEFFAKRRLVTIFS 895
Query: 266 APNYCYRCGNMASILE 281
APNY N ++E
Sbjct: 896 APNYGGEFDNARCVIE 943
>TC86910 homologue to PIR|T09548|T09548 phosphoprotein phosphatase (EC
3.1.3.16) 1 catalytic chain delta - alfalfa, complete
Length = 1523
Score = 217 bits (553), Expect(2) = 2e-71
Identities = 99/189 (52%), Positives = 134/189 (70%), Gaps = 1/189 (0%)
Frame = +2
Query: 27 LSEQQVKELCEKAKEILMEESNVQPVKSPVTICGDIHGQFHDLAELFRIGGKCPDTNYLF 86
LSE ++K+LC +K+I M + N+ +++P+ ICGDIHGQ+ DL LF GG P +NYLF
Sbjct: 320 LSEAEIKQLCLVSKDIFMNQPNLLKLEAPIKICGDIHGQYSDLLRLFEYGGFPPRSNYLF 499
Query: 87 MGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHESRQITQVYGFYDECLRKYGNANV 146
+GDYVDRG S+ET+ LL+A K++Y + +LRGNHE I ++YGFYDEC R+Y N +
Sbjct: 500 LGDYVDRGKQSLETICLLLAYKIKYPKNFFLLRGNHECASINRIYGFYDECKRRY-NVKL 676
Query: 147 WKTFTDLFDFFPLTALVESEIFCLHGGLSPSIETLDNIRNFDRVQEVPHEGPMCDLLWSD 206
WK FTD F+ P+ AL++ +I C+HGGLSP + L+ I+N R EVP G +CDLLWSD
Sbjct: 677 WKMFTDCFNCLPVAALIDEKIICMHGGLSPELHDLNQIKNLRRPCEVPESGLLCDLLWSD 856
Query: 207 P-DDRCGWG 214
P D GWG
Sbjct: 857 PSSDVRGWG 883
Score = 69.7 bits (169), Expect(2) = 2e-71
Identities = 35/88 (39%), Positives = 54/88 (60%)
Frame = +3
Query: 212 GWGISPRGAGYTFGQDISEQFNHTNSLKLIARAHQLVMDGFNWAHEQKVVTIFSAPNYCY 271
G G S RG YTFG D ++F + L LI RAHQ+V DG+ + +++VTIFSAPNYC
Sbjct: 876 GGGESERGVSYTFGADRVKEFLQKHDLDLICRAHQVVEDGYEFFANRQLVTIFSAPNYCG 1055
Query: 272 RCGNMASILEVDDCKGHTFIQFEPAPRR 299
N+ +++ V++ +F +P ++
Sbjct: 1056DFDNVGAMMTVNESLVCSFQILKPLDKK 1139
>TC81378 homologue to GP|19071628|gb|AAL84295.1 serine/threonine protein
phosphatase PP2A-4 catalytic subunit, partial (28%)
Length = 768
Score = 206 bits (524), Expect = 9e-54
Identities = 94/98 (95%), Positives = 97/98 (98%)
Frame = +3
Query: 216 SPRGAGYTFGQDISEQFNHTNSLKLIARAHQLVMDGFNWAHEQKVVTIFSAPNYCYRCGN 275
SPRGAGYTFGQDISEQFNHTN+LKLIARAHQLVM+G+NW HEQKVVTIFSAPNYCYRCGN
Sbjct: 12 SPRGAGYTFGQDISEQFNHTNNLKLIARAHQLVMEGYNWGHEQKVVTIFSAPNYCYRCGN 191
Query: 276 MASILEVDDCKGHTFIQFEPAPRRGEPDVTRRTPDYFL 313
MASILEVDDCKGHTFIQFEPAPRRGEPDVTRRTPDYFL
Sbjct: 192 MASILEVDDCKGHTFIQFEPAPRRGEPDVTRRTPDYFL 305
>TC87324 homologue to PIR|E86217|E86217 protein T27G7.10 [imported] -
Arabidopsis thaliana, partial (37%)
Length = 1897
Score = 198 bits (503), Expect = 3e-51
Identities = 109/265 (41%), Positives = 154/265 (57%), Gaps = 15/265 (5%)
Frame = +2
Query: 31 QVKELCEKAKEILMEESNVQPVKSPVTICGDIHGQFHDLAELFRIGGKCPDT-------N 83
++ +LC+ A+ I E V +++P+ I GD+HGQF DL LF G P T +
Sbjct: 314 EIADLCDSAERIFSSEPTVIQLRAPIKIFGDLHGQFGDLMRLFDEYGS-PSTAGDIAYID 490
Query: 84 YLFMGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHESRQITQVYGFYDECLRKYGN 143
YLF+GDYVDRG +S+ET++LL+ALKV Y I ++RGNHE+ I ++GF EC+ + G
Sbjct: 491 YLFLGDYVDRGQHSLETISLLLALKVEYPNNIHLIRGNHEAADINALFGFRIECIERMGE 670
Query: 144 AN---VWKTFTDLFDFFPLTALVESEIFCLHGGLSPSIETLDNIRNFDR-VQEVPHEGPM 199
+ W LF++ PL AL+E +I C+HGG+ SI L+ I N R + +
Sbjct: 671 RDGIWTWHRINRLFNWLPLAALIEKKIICMHGGIGRSINHLEQIENIQRPITMEAGSIVL 850
Query: 200 CDLLWSDPDDRCG-WGISPRGAG---YTFGQDISEQFNHTNSLKLIARAHQLVMDGFNWA 255
DLLWSDP + G+ P G TFG D +F + N L+LI RAH+ VMDGF
Sbjct: 851 MDLLWSDPTENDSVEGLRPNARGPGLVTFGPDRVMEFCNNNDLQLIVRAHECVMDGFERF 1030
Query: 256 HEQKVVTIFSAPNYCYRCGNMASIL 280
+ ++T+FSA NYC N +IL
Sbjct: 1031AQGHLITLFSATNYCGTANNAGAIL 1105
>TC89103 similar to GP|16930441|gb|AAL31906.1 At2g42810/F7D19.19
{Arabidopsis thaliana}, partial (66%)
Length = 1107
Score = 162 bits (411), Expect(2) = 3e-42
Identities = 97/244 (39%), Positives = 140/244 (56%), Gaps = 5/244 (2%)
Frame = +1
Query: 66 FHDLAELFRIGGKCPDTN-YLFMGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHES 124
F+DL +F + G + N YLF GD+VDRG +SVE L A K + I RGNHES
Sbjct: 250 FYDLLNIFELNGIPSEENPYLFNGDFVDRGSFSVEVAFTLFAFKCMSPSAMYIARGNHES 429
Query: 125 RQITQVYGFYDECLRKYGNANVWKTFTDLFDFFPLTALVESEIFCLHGGL-SPSIETLDN 183
+ + +++GF E K + V K F ++F+ PL ++ ++F +HGGL S L
Sbjct: 430 KSMNKIFGFEGEVRAKLNDKFV-KMFAEVFNCLPLAHVINKKVFVVHGGLFSVDGVKLSQ 606
Query: 184 IRNFDRVQEVPHEGPMCDLLWSDPDDRCGWGISPRGAG-YTFGQDISEQFNHTNSLKLIA 242
IR+ +R E P EG M DLLWSDP G G S RG + FG+D++++F N+L L+
Sbjct: 607 IRSINRFCEPPDEGLMHDLLWSDPQPLPGRGPSKRGNRCFCFGEDVTKRFLKDNNLDLVV 786
Query: 243 RAHQLVMDGFNWAHEQKVVTIFSAPNYCYRCGNMASIL--EVDDCKGHTFIQFEPAPRRG 300
R+H+ + G+ H+ K++T+FSAPNYC + GN + + E D K + I FE P
Sbjct: 787 RSHKSMHKGYEIQHDGKLITVFSAPNYCDKKGNKGAFIRFEAPDLKPN-IITFEAVP--- 954
Query: 301 EPDV 304
PDV
Sbjct: 955 HPDV 966
Score = 26.9 bits (58), Expect(2) = 3e-42
Identities = 14/54 (25%), Positives = 27/54 (49%), Gaps = 4/54 (7%)
Frame = +3
Query: 16 EQISQLMQCKPLSEQQVKELCEKAKEILMEESNVQPVKSP----VTICGDIHGQ 65
+ ++ + K L ++ + K+IL ++ + P VT+CGD+HGQ
Sbjct: 81 KMMNDFKEQKYLDRGYASKILLQTKKILKALPSLVDITVPHGKHVTVCGDVHGQ 242
>AL378129 GP|20385063|g serine/threonine protein phosphatase 2A {Pisum
sativum}, partial (37%)
Length = 421
Score = 160 bits (406), Expect = 5e-40
Identities = 75/115 (65%), Positives = 93/115 (80%)
Frame = +2
Query: 18 ISQLMQCKPLSEQQVKELCEKAKEILMEESNVQPVKSPVTICGDIHGQFHDLAELFRIGG 77
IS++ + L E +++ LCE KEIL+EESNVQPV SPVT+CGDIHGQFHDL +LF+ GG
Sbjct: 77 ISKVKDGQHLLEDELQLLCEYVKEILIEESNVQPVNSPVTVCGDIHGQFHDLMKLFQTGG 256
Query: 78 KCPDTNYLFMGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHESRQITQVYG 132
P+TNY+FMGD+VDRGY S+E T+L+ LK RY IT+LRGNHESRQ+TQVYG
Sbjct: 257 HVPETNYIFMGDFVDRGYNSLEVFTILLLLKARYPANITLLRGNHESRQLTQVYG 421
>AW686729 GP|7248361|dbj type 2A protein phosphatase-2 {Vicia faba}, partial
(29%)
Length = 529
Score = 147 bits (370), Expect = 7e-36
Identities = 64/90 (71%), Positives = 77/90 (85%)
Frame = +3
Query: 224 FGQDISEQFNHTNSLKLIARAHQLVMDGFNWAHEQKVVTIFSAPNYCYRCGNMASILEVD 283
FGQDI+ QFNHTN L LI+RAHQLVM+G+NWA E+ VVT+FSAPNYCYRCGNMA+ILE+
Sbjct: 3 FGQDIASQFNHTNGLSLISRAHQLVMEGYNWAQEKNVVTVFSAPNYCYRCGNMAAILEIG 182
Query: 284 DCKGHTFIQFEPAPRRGEPDVTRRTPDYFL 313
+ F+QF+PAPR+ EPD TR+TPDYFL
Sbjct: 183 ENMDQNFLQFDPAPRQIEPDTTRKTPDYFL 272
>TC79344 homologue to PIR|E86217|E86217 protein T27G7.10 [imported] -
Arabidopsis thaliana, partial (22%)
Length = 1301
Score = 139 bits (349), Expect = 2e-33
Identities = 76/190 (40%), Positives = 108/190 (56%), Gaps = 10/190 (5%)
Frame = +1
Query: 101 VTLLVALKVRYHQRITILRGNHESRQITQVYGFYDECLRKYGNAN---VWKTFTDLFDFF 157
+TLL+ALKV Y + ++RGNHE+ I ++GF EC+ + G + W LF++
Sbjct: 7 ITLLLALKVEYPTNVHLIRGNHEAADINALFGFRIECIERMGERDGIWTWHRINRLFNWL 186
Query: 158 PLTALVESEIFCLHGGLSPSIETLDNIRNFDRVQEVPHEGP---MCDLLWSDPDDRCGW- 213
PL AL+E +I C+HGG+ SI ++ I N R +P E + DLLWSDP +
Sbjct: 187 PLAALIEKKIICMHGGIGRSINHIEQIENIQR--PIPMEAGSIVLMDLLWSDPTENDSVE 360
Query: 214 GISPRGAG---YTFGQDISEQFNHTNSLKLIARAHQLVMDGFNWAHEQKVVTIFSAPNYC 270
G+ P G TFG D +F + N L+LI RAH+ VMDGF + ++T+FSA NYC
Sbjct: 361 GLRPNARGPGLVTFGPDRVMEFCNNNDLQLIVRAHECVMDGFERFAQGHLITLFSATNYC 540
Query: 271 YRCGNMASIL 280
N +IL
Sbjct: 541 GTANNAGAIL 570
>TC89568 homologue to PIR|T09544|T09544 phosphoprotein phosphatase (EC
3.1.3.16) catalytic beta chain - alfalfa, partial (52%)
Length = 807
Score = 139 bits (349), Expect = 2e-33
Identities = 66/142 (46%), Positives = 93/142 (65%), Gaps = 1/142 (0%)
Frame = +3
Query: 159 LTALVESEIFCLHGGLSPSIETLDNIRNFDRVQEVPHEGPMCDLLWSDPD-DRCGWGISP 217
+ AL++ +IFC+HGGLSP + LD IRN R +VP G +CDLLWSDP D GWG++
Sbjct: 3 VAALIDEKIFCMHGGLSPDLHNLDQIRNLQRPTDVPDTGLLCDLLWSDPSKDVQGWGMND 182
Query: 218 RGAGYTFGQDISEQFNHTNSLKLIARAHQLVMDGFNWAHEQKVVTIFSAPNYCYRCGNMA 277
RG YTFG D +F + L LI RAHQ+V DG+ + ++++VTIFSAPNYC N
Sbjct: 183 RGVSYTFGADKITEFLEKHDLDLICRAHQVVEDGYEFFADRQLVTIFSAPNYCGEFDNAG 362
Query: 278 SILEVDDCKGHTFIQFEPAPRR 299
+++ VD+ +F +PA ++
Sbjct: 363 AMMSVDETLMCSFQILKPADKK 428
>TC90362 homologue to PIR|T09547|T09547 phosphoprotein phosphatase (EC
3.1.3.16) 1 catalytic gsmma chain - alfalfa, partial
(39%)
Length = 617
Score = 124 bits (312), Expect = 4e-29
Identities = 55/128 (42%), Positives = 89/128 (68%), Gaps = 7/128 (5%)
Frame = +3
Query: 14 LDEQISQLMQCK-------PLSEQQVKELCEKAKEILMEESNVQPVKSPVTICGDIHGQF 66
+D+ I++L++ + LSE ++++LC +++I +++ + +++P+ ICGDIHGQ+
Sbjct: 213 VDDIINRLLEVRNRPGKQVQLSESEIRQLCNVSRDIFLKQPFLLQLEAPIKICGDIHGQY 392
Query: 67 HDLAELFRIGGKCPDTNYLFMGDYVDRGYYSVETVTLLVALKVRYHQRITILRGNHESRQ 126
DL LF GG P++NYLF+GDYVDRG S+ET+ LL+A K++Y +LRGNHE
Sbjct: 393 SDLLRLFEYGGLPPESNYLFLGDYVDRGKQSLETICLLLAYKIKYPDNFFLLRGNHECAS 572
Query: 127 ITQVYGFY 134
I ++YGFY
Sbjct: 573 INRIYGFY 596
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.139 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,800,001
Number of Sequences: 36976
Number of extensions: 159832
Number of successful extensions: 629
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 602
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 607
length of query: 313
length of database: 9,014,727
effective HSP length: 96
effective length of query: 217
effective length of database: 5,465,031
effective search space: 1185911727
effective search space used: 1185911727
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0029b.5


