

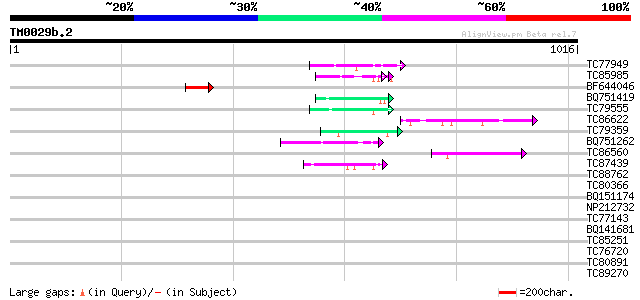
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0029b.2
(1016 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77949 weakly similar to GP|11762218|gb|AAG40387.1 AT4g27520 {A... 50 4e-06
TC85985 similar to GP|5139695|dbj|BAA81686.1 expressed in cucumb... 47 3e-05
BF644046 45 1e-04
BQ751419 weakly similar to GP|21629340|gb L509.2 {Leishmania maj... 45 1e-04
TC79555 homologue to GP|10567859|gb|AAG18593.1 L8019.5 {Leishman... 44 3e-04
TC86622 weakly similar to GP|20197623|gb|AAM15156.1 putative myo... 44 4e-04
TC79359 weakly similar to PIR|S49915|S49915 extensin-like protei... 44 4e-04
BQ751262 similar to GP|6715100|gb|A versicolorin B synthase {Asp... 43 5e-04
TC86560 similar to GP|8843737|dbj|BAA97285.1 myosin heavy chain-... 43 5e-04
TC87439 weakly similar to GP|5139695|dbj|BAA81686.1 expressed in... 43 7e-04
TC88762 similar to GP|22136928|gb|AAM91808.1 unknown protein {Ar... 42 0.002
TC80366 similar to GP|13877617|gb|AAK43886.1 protein kinase-like... 41 0.003
BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xy... 41 0.003
NP212732 NP212732|AF106929.1|AAD39890.1 putative cell wall protein 40 0.003
TC77143 similar to GP|559557|gb|AAA51649.1|| arabinogalactan-pro... 40 0.005
BQ141681 weakly similar to GP|15145793|gb| basic proline-rich pr... 39 0.008
TC85251 homologue to PIR|JQ2128|JQ2128 metallothionein - soybean... 39 0.008
TC76720 similar to GP|11121502|emb|CAC14888. putative extensin {... 39 0.010
TC80891 similar to GP|21622328|emb|CAD36969. probable isoamyl al... 39 0.010
TC89270 similar to GP|22773423|gb|AAL66352.2 catalase-peroxidase... 38 0.017
>TC77949 weakly similar to GP|11762218|gb|AAG40387.1 AT4g27520 {Arabidopsis
thaliana}, partial (34%)
Length = 1297
Score = 50.1 bits (118), Expect = 4e-06
Identities = 49/178 (27%), Positives = 75/178 (41%), Gaps = 6/178 (3%)
Frame = +3
Query: 538 APKRRRLTKTSSGAGTSNPGAQPTTAA-APKGKNVAEASIAAATEPTAVPASTPASAGAT 596
+P+ + + + S G S P PTT +P G + +I A+ P VP S P ++ +
Sbjct: 486 SPRTPKSSPSPSAGGLSPPSPSPTTTTPSPSGSPPSPVAIPPASSP--VPTSGPTASSPS 659
Query: 597 AAVAAESSVGATAASAG-VNATKAA----ASSDTPIGEKEKENETPKSPPHQEAPPSPPP 651
V+ + G A+S V +T A ASS +P G + PP+P P
Sbjct: 660 PVVSTPPAGGPMASSPSPVVSTPPAGGPMASSPSPAGGPPALSPAGGPSTAAGGPPAPGP 839
Query: 652 TRDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSSSYYNMLPNAIEPSEFLL 709
A + P P G S PG A + + + AP S S+ + PS FL+
Sbjct: 840 G-GAATSPGPGGAGA-SGPGGAAAAPGSPGSNSTAPSGNSGSF-------VAPSNFLV 986
>TC85985 similar to GP|5139695|dbj|BAA81686.1 expressed in cucumber
hypocotyls {Cucumis sativus}, partial (42%)
Length = 892
Score = 47.4 bits (111), Expect = 3e-05
Identities = 42/131 (32%), Positives = 58/131 (44%), Gaps = 5/131 (3%)
Frame = +3
Query: 549 SGAGTSNPGAQPTTAAAPKGKNVAEASIAAATEPTAVPASTPASAGATAAVAAESSVGAT 608
S A +S A +T P + A + ++ PT VP S+P +A +T AV + V A
Sbjct: 315 SPAASSPVVAPVSTPPKPAPVSSPPAPVPVSSPPTPVPVSSPPTA-STPAVTPSAEVPAA 491
Query: 609 AASAGVNATKAAASSDTPIGEKEKENETPKSPPHQEAPPSPP-----PTRDAGSMPSPPH 663
A S TK K K++ P P E PP+PP P+ DA S P P
Sbjct: 492 APSKSKKKTK-----------KGKKHSAPAPSPALEGPPAPPVGAPGPSLDASS-PGPAS 635
Query: 664 QGEKSCPGAAT 674
++S GA T
Sbjct: 636 AADES--GAET 662
Score = 45.4 bits (106), Expect = 1e-04
Identities = 42/152 (27%), Positives = 66/152 (42%), Gaps = 13/152 (8%)
Frame = +3
Query: 549 SGAGTSNPGAQPTTAAAPKGKNVAEASIAAATEPTA-VPASTPASAGATAAVAAESSVGA 607
+G G +P + PTT+ P + A +AA T+P + P ++P S+ ++ A + A
Sbjct: 105 AGVGGQSPSSAPTTSP-PTVTTPSAAPVAAPTKPKSPAPVASPKSSPPASSPTAATVTPA 281
Query: 608 TAASAGVNATKAAASSD---TPIGEKEKENETPKSPPHQEAPPSPPPTRDAGSMP----S 660
+ +A V K+ A+S P+ K P S P P S PPT S P +
Sbjct: 282 VSPAAPVPVAKSPAASSPVVAPVSTPPKP--APVSSPPAPVPVSSPPTPVPVSSPPTAST 455
Query: 661 PPHQGEKSCPGAATTSEAAQIEQ-----APAP 687
P P AA + + ++ APAP
Sbjct: 456 PAVTPSAEVPAAAPSKSKKKTKKGKKHSAPAP 551
>BF644046
Length = 597
Score = 45.4 bits (106), Expect = 1e-04
Identities = 20/51 (39%), Positives = 31/51 (60%)
Frame = +3
Query: 315 YEYMFSELGIRLPFSPFVQTVLRDINAAPCQLHPNAWAFIRCFEILSAAVG 365
Y ++F ++G + PF+ F L+ +N A QLHPN AF+ FEI ++G
Sbjct: 15 YSFVFEDIGFKFPFTNFECDFLKALNVASSQLHPNCCAFMCGFEISCESLG 167
>BQ751419 weakly similar to GP|21629340|gb L509.2 {Leishmania major}, partial
(1%)
Length = 766
Score = 45.1 bits (105), Expect = 1e-04
Identities = 41/153 (26%), Positives = 56/153 (35%), Gaps = 13/153 (8%)
Frame = +1
Query: 548 SSGAGTSNPGAQPTTAAA-PKGKNVAEASIAAATEPTAVPASTPASAGATAAVAAESSVG 606
+S + ++ P PTTA++ P G A T + +TPA A TA +
Sbjct: 52 ASSSPSARPSPPPTTASSTPPG---APTPSPTPTRRATLNGNTPAPASPTAQATPPTGPS 222
Query: 607 ATAASAGVNATKAAASSDTPIGEKEKENETPKSP-PHQE-APPSPPPTRDAGSMPSPP-- 662
A S T SS T + SP PH PP+P P+ S PP
Sbjct: 223 TAAPSPSTCTTTGPTSSSTSASSPAATSPRTTSPSPHSSGTPPAPAPSASRSSTSRPPRA 402
Query: 663 ---HQGEKSCP-----GAATTSEAAQIEQAPAP 687
+S P A TT+ + Q P P
Sbjct: 403 RARRAASRSSPWATPARACTTAPTSASRQTPPP 501
Score = 34.7 bits (78), Expect = 0.19
Identities = 28/109 (25%), Positives = 37/109 (33%)
Frame = +1
Query: 518 NTEGDAKKRGGAENQAESTKAPKRRRLTKTSSGAGTSNPGAQPTTAAAPKGKNVAEASIA 577
+T + R S A R T + A P P TAA PK +
Sbjct: 379 STSRPPRARARRAASRSSPWATPARACTTAPTSASRQTPPPSPRTAAKPKASRCTPSRSR 558
Query: 578 AATEPTAVPASTPASAGATAAVAAESSVGATAASAGVNATKAAASSDTP 626
A TEP S + A AA+AG AT+ A++ P
Sbjct: 559 APTEP--------------------SKMAAAAAAAGTRATRPRATAAAP 645
Score = 32.0 bits (71), Expect = 1.2
Identities = 25/98 (25%), Positives = 37/98 (37%), Gaps = 3/98 (3%)
Frame = +1
Query: 610 ASAGVNATKAAASSDTPIGEKEKENETPKSPPHQEAPPSPPPTRDA---GSMPSPPHQGE 666
A++ ++ T +S +P T S P PSP PTR A G+ P+P
Sbjct: 16 AASNLHTTWQPPASSSPSARPSPPPTTASSTPPGAPTPSPTPTRRATLNGNTPAPASPTA 195
Query: 667 KSCPGAATTSEAAQIEQAPAPEVGSSSYYNMLPNAIEP 704
++ P ++ A SSS P A P
Sbjct: 196 QATPPTGPSTAAPSPSTCTTTGPTSSSTSASSPAATSP 309
>TC79555 homologue to GP|10567859|gb|AAG18593.1 L8019.5 {Leishmania major},
partial (1%)
Length = 1410
Score = 43.9 bits (102), Expect = 3e-04
Identities = 43/165 (26%), Positives = 63/165 (38%), Gaps = 14/165 (8%)
Frame = +2
Query: 537 KAPKRRRLTKTSSGAGTSNPGAQPTTAAAPKGKNVAEASIAAATEPTAVPASTPASAGAT 596
K+P R R + S S G +P AAA G + A+ PT ++ A G
Sbjct: 722 KSPPRPRPARLPSARRASPRGGRPLAAAAAAGATTS----TTASRPTRSARTSAARRGRP 889
Query: 597 AAVAAESSVGATAASAGVNATKAAASSDTPIGEKEKENETPKSPPHQEAPPSP------- 649
A S G+ G + T+A + + + + + S P P SP
Sbjct: 890 AI*DGLCSSGSMRTCRG*SRTRALTFTSSTVSVRSNGSFGASSGPSNTCPSSPRRRSGRL 1069
Query: 650 ------PPTRDAGSMPSPPHQGEKSCPGA-ATTSEAAQIEQAPAP 687
P +A S P PP S P A TS++A ++PAP
Sbjct: 1070PAATTSSPATNANSPPFPPASPSSSVPPAKPPTSKSA---RSPAP 1195
Score = 36.2 bits (82), Expect = 0.066
Identities = 33/114 (28%), Positives = 53/114 (45%), Gaps = 9/114 (7%)
Frame = +2
Query: 519 TEGDAKKRGGAENQAESTKAPKRRRL--TKTSSGAGTSN----PGAQPTTAAAPKGKNVA 572
+ G G N S+ + RL TSS A +N P A P+++ P +
Sbjct: 992 SNGSFGASSGPSNTCPSSPRRRSGRLPAATTSSPATNANSPPFPPASPSSSVPPAKPPTS 1171
Query: 573 EASIAAA---TEPTAVPASTPASAGATAAVAAESSVGATAASAGVNATKAAASS 623
+++ + A + P+ P+S+P SA A+A+ +S A SAG T+ ASS
Sbjct: 1172 KSARSPAPV*SRPSPPPSSSPGSA-ASASSRCSASSRARPTSAGSRRTRTTASS 1330
Score = 34.3 bits (77), Expect = 0.25
Identities = 32/124 (25%), Positives = 47/124 (37%), Gaps = 11/124 (8%)
Frame = +2
Query: 581 EPTAVPASTPASAGATAAVAAESSVGATAASAGVNATKAAASSDTPI-----GEKEKENE 635
+PT VP +P + ++ A S + A + T + + TP +
Sbjct: 455 KPTLVPP*SPQPSSTRSSPAPPSRPSSPATTTRPPPTSKPSPTQTPSRSPSSARTPSTSS 634
Query: 636 TPKSPPHQEAPPSPP---PTRDAG--SMP-SPPHQGEKSCPGAATTSEAAQIEQAPAPEV 689
TP P +PP PP P R G S+P SPP P A S A A
Sbjct: 635 TPTRAPLLSSPPPPPPFFPARRPGTRSLPKSPPRPRPARLPSARRASPRGGRPLAAAAAA 814
Query: 690 GSSS 693
G+++
Sbjct: 815 GATT 826
>TC86622 weakly similar to GP|20197623|gb|AAM15156.1 putative myosin heavy
chain {Arabidopsis thaliana}, partial (17%)
Length = 2546
Score = 43.5 bits (101), Expect = 4e-04
Identities = 67/285 (23%), Positives = 116/285 (40%), Gaps = 39/285 (13%)
Frame = +2
Query: 700 NAIEPSEFLLAGLNRDV-------IEKEVLSRGLNDTKEETLACLLRAGCIFAHTFEKFN 752
NAIE + LA RD+ +E E L L E+ L EKFN
Sbjct: 362 NAIEEN---LAVRGRDIELTSARNLELESLHESLTRDSEQKLQ----------EAIEKFN 502
Query: 753 AANVEAERLKAESAKHQEAAAA-----------WEKRFDKLATQAGKDK------VYADK 795
+ + E + L + +E A +E+ KLA+ +++ V A+K
Sbjct: 503 SKDSEVQSLLEKIKILEENIAGAGEQSISLKSEFEESLSKLASLQSENEDLKRQIVEAEK 682
Query: 796 MIGTAGIKIGELEDQLALMKEEADELDASLQACKKEKEQAEKDLIARGEAL--------- 846
+ + L +K + DEL SL + EKE ++L++ L
Sbjct: 683 KTSQSFSENELLVGTNIQLKTKIDELQESLNSVVSEKEVTAQELVSHKNLLAELNDVQSK 862
Query: 847 ---IAKESELAVLCAELELVKKALAEQEKKSAESLALAKSDMEAVMQATSEEIKKATETH 903
I +E+ +L E +L ++AL + +K +E+ ++ + +IK E
Sbjct: 863 SSEIHSANEVRILEVESKL-QEALQKHTEKESET-----KELNEKLNTLEGQIKIYEEQA 1024
Query: 904 AEALA---TKDAEIASQLAKIKSLEDELATEKAKAIEAREQAADI 945
EA+A + AE+ L K+K LE + ++ K++E + A I
Sbjct: 1025HEAVAAAENRKAELEESLIKLKHLEAAVEEQQNKSLERETETAGI 1159
Score = 34.3 bits (77), Expect = 0.25
Identities = 36/152 (23%), Positives = 63/152 (40%), Gaps = 33/152 (21%)
Frame = +3
Query: 756 VEAERLKAESAKHQEAAAAWEKRFDKLATQAGKDKVYADKM--IGTAGIKIGELEDQLAL 813
+EA+ KAES H+E + + + K + Y K+ I K+ ELE +L L
Sbjct: 1548 IEAQLAKAESRLHEEVGSVQAAASQREVDLSSKFEDYEQKISEINVLNGKVAELEKELHL 1727
Query: 814 MK--------EEAD--ELDASLQACKKEKE---------------------QAEKDLIAR 842
+ EE+ EL+A+L+ +E E QA++ + +
Sbjct: 1728 AQDTIANQKGEESQKLELEAALKNSVEELETKKNEISLLQKQVIEFEQKLQQADEKISVK 1907
Query: 843 GEALIAKESELAVLCAELELVKKALAEQEKKS 874
GE + K+ L V + + + + +KKS
Sbjct: 1908 GEEAVDKKDALEVKSRDFSISSPSKRKSKKKS 2003
>TC79359 weakly similar to PIR|S49915|S49915 extensin-like protein - maize,
partial (3%)
Length = 1170
Score = 43.5 bits (101), Expect = 4e-04
Identities = 39/156 (25%), Positives = 57/156 (36%), Gaps = 9/156 (5%)
Frame = +3
Query: 558 AQPTTAAAPKGKNVAEASIAAATEPTAVPAS-----TPASAGATAAVAAESSVGATAASA 612
AQ A++PK A+ + + PT PAS T + T AV+ S +
Sbjct: 216 AQNAPASSPKSSVTAKPPSSVSVSPTNSPASPAKSPTLSPPSQTPAVSPSGSASTPPPAT 395
Query: 613 GVNATKAAASSDTPIGEKEKENETPKSPPHQEAPPSPPPTRDAGSMPSPPHQGEKSCPGA 672
A A + + + S P +P S PPT + SP S P
Sbjct: 396 SPPAKSPAVQPPSSVSPAISPSNNVSSTPPVSSPASSPPTAAVSPVSSPVEAPSVSSPPE 575
Query: 673 ATT----SEAAQIEQAPAPEVGSSSYYNMLPNAIEP 704
A++ S +A APA + SS P + P
Sbjct: 576 ASSAGIPSSSATPADAPAATLPSSKSPGTSPASSSP 683
>BQ751262 similar to GP|6715100|gb|A versicolorin B synthase {Aspergillus
parasiticus}, partial (11%)
Length = 710
Score = 43.1 bits (100), Expect = 5e-04
Identities = 45/187 (24%), Positives = 78/187 (41%), Gaps = 2/187 (1%)
Frame = +3
Query: 486 SRTPKYLEDMNFTNAELLKARERRMARFSPKFNTEGDAKKRGGAENQAESTKAPKRRRL- 544
+R+ + L T+ L RR + P+ + + + R R+
Sbjct: 162 TRSGRTLSATTLTHGTLSSPTSRRASSSLPRVLRDSPTPAPSTTQMPSPPPAGLLRSRMP 341
Query: 545 TKTSSGAGTSNPGAQPTTAAAPKGK-NVAEASIAAATEPTAVPASTPASAGATAAVAAES 603
T S A S+P + T +A+P+ + + AE+S A +T PT P +++ A + +
Sbjct: 342 TMPSRSAHISSPLS--TRSASPRPRTSTAESSWARSTAPTPSPQRRKSASRARRRSSPKP 515
Query: 604 SVGATAASAGVNATKAAASSDTPIGEKEKENETPKSPPHQEAPPSPPPTRDAGSMPSPPH 663
S T+ S + + ++S+ T K SPP +PP+ P+R A SPP
Sbjct: 516 SAARTSRSTPSPSPRRSSSTAT------KRPPASSSPPTPASPPT--PSRRARRSSSPPA 671
Query: 664 QGEKSCP 670
E P
Sbjct: 672 PSEPPAP 692
>TC86560 similar to GP|8843737|dbj|BAA97285.1 myosin heavy chain-like
{Arabidopsis thaliana}, partial (28%)
Length = 2450
Score = 43.1 bits (100), Expect = 5e-04
Identities = 52/191 (27%), Positives = 84/191 (43%), Gaps = 21/191 (10%)
Frame = +2
Query: 756 VEAERLKAESAKHQEAAAAWEKRFDKL------------ATQAGKDKVY--ADKMIGTAG 801
VE E +K E ++ +E + E L A A + KV +++MI T
Sbjct: 1301 VELENVKKEHSELKEKESELESTVGNLHVKLRKSKSELEACSADESKVRGASEEMILTLS 1480
Query: 802 IKIGELED---QLALMKEEADELDASLQACKKEKEQAEKDL-IARGEALIAKESELAVLC 857
E E+ ++ MK + DEL +A K E+AEK L A EA AK +E + +
Sbjct: 1481 RLTSETEEARREVEDMKNKTDELKKEAEATKLALEEAEKKLKEATEEAEAAKAAEASAIE 1660
Query: 858 AELELVKKALAEQEKKSAES---LALAKSDMEAVMQATSEEIKKATETHAEALATKDAEI 914
L ++ A + S+ES + ++ + E++ + E K A A A +A
Sbjct: 1661 QITVLTERTSARRASTSSESGAAMTISTEEFESLKRKVEESDKLADMKVDAAKAQVEAVK 1840
Query: 915 ASQLAKIKSLE 925
AS+ +K LE
Sbjct: 1841 ASENEVLKKLE 1873
Score = 38.5 bits (88), Expect = 0.013
Identities = 67/314 (21%), Positives = 116/314 (36%), Gaps = 22/314 (7%)
Frame = +2
Query: 647 PSPPPTRDAGSMP-SPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSSSYYNMLPNAIEPS 705
PSP + G + SPP Q K + E A + P + P A
Sbjct: 329 PSPSLKPEVGEIDTSPPFQSVKDA--VSLFGEGAFSGERPI---------HKKPKAYSAE 475
Query: 706 EFLLAGLNRDVIEKEVLSRGLNDTKEETLACLLRAGCIFAHTFEKFNAANVEAERLKAES 765
L V EKE LN KE+ + ++LK +
Sbjct: 476 RVLAKETQLHVAEKE-----LNKLKEQVKNAETTKAQALVELERAQRTVDDLTQKLKLIT 640
Query: 766 AKHQEAAAAWEKRFDKLATQAGKDKVYADKMIGTAGIKIGELEDQLALMKEEADELDASL 825
+ A A E + + G+ G G ELE+ + ELDA+
Sbjct: 641 ESRESAVKATEAAKSQAKQKYGESD-------GVNGAWKEELENAVQRYASIMTELDAAK 799
Query: 826 QACKKEKEQAEKDLIARGEALIAKE----------SELAVLCAELELVKKA-----LAEQ 870
Q +K +++ + AR A+ E ++ L E+ VK++ LA
Sbjct: 800 QDLRKTRQEYDSSSDARVSAVKRTEEAENAMKENTERVSELSKEISAVKESIEQTKLAYV 979
Query: 871 EKKSAESLALAKSD-MEAVMQATSEEIKK-----ATETHAEALATKDAEIASQLAKIKSL 924
E + ++L L + D + +AT E+ KK E + E + +A++A + +I +L
Sbjct: 980 ESQQQQALVLTEKDALRQSYKATLEQSKKKLLALKKEFNPEITKSLEAQLAETMNEIAAL 1159
Query: 925 EDELATEKAKAIEA 938
EL +++ +++
Sbjct: 1160HTELENKRSSDLDS 1201
>TC87439 weakly similar to GP|5139695|dbj|BAA81686.1 expressed in cucumber
hypocotyls {Cucumis sativus}, partial (57%)
Length = 897
Score = 42.7 bits (99), Expect = 7e-04
Identities = 48/166 (28%), Positives = 69/166 (40%), Gaps = 16/166 (9%)
Frame = +3
Query: 527 GGAENQAESTKAPKRRRLTKTSSGAGTSNPGAQPTTAAAPKGKNVAEASIAAATEPTAVP 586
GG A T +P T ++ +S A PTT P ++S A + A P
Sbjct: 96 GGQSPSAAPTTSPS----TPAATTPVSSPVAAPPTTPTTPAPVASPKSSPPATSPKAAAP 263
Query: 587 ASTPASAGATAAVAAESS-------VGATAASAGVNA----TKAAASSDTPIGEKEKENE 635
+TP +A + AV S+ V + A V++ T AA + TP ++
Sbjct: 264 TATPPAASSPPAVTPVSTPPPAPVPVKSPPTPAPVSSPPAVTPVAAPTTTPAVPAPAPSK 443
Query: 636 TPKSPPHQEAP-PSP----PPTRDAGSMPSPPHQGEKSCPGAATTS 676
K+ AP PSP PP AG+ P P + S PG ATT+
Sbjct: 444 GKKNKKKHGAPAPSPALLGPPAPPAGA-PGPSE--DASSPGPATTA 572
Score = 41.6 bits (96), Expect = 0.002
Identities = 47/166 (28%), Positives = 56/166 (33%), Gaps = 5/166 (3%)
Frame = +3
Query: 549 SGAGTSNPGAQPTTAAAPKGKNVAEASIAAATEPTAVPASTPASAGATAAVAAESSVGAT 608
+G G +P A PTT+ S AAT P + P + P + T A A
Sbjct: 87 AGVGGQSPSAAPTTSP----------STPAATTPVSSPVAAPPTTPTTPAPVASPKSSPP 236
Query: 609 AASAGVNATKAAASSDTPIGEKEKENETPKSPPHQEAPPSPPPTRDAGSMP----SPPHQ 664
A S KAAA + TP P +PP+ P P SPP
Sbjct: 237 ATSP-----KAAAPTATP--------------PAASSPPAVTPVSTPPPAPVPVKSPPTP 359
Query: 665 GEKSCPGAATTSEAAQIEQA-PAPEVGSSSYYNMLPNAIEPSEFLL 709
S P A T A A PAP A PS LL
Sbjct: 360 APVSSPPAVTPVAAPTTTPAVPAPAPSKGKKNKKKHGAPAPSPALL 497
>TC88762 similar to GP|22136928|gb|AAM91808.1 unknown protein {Arabidopsis
thaliana}, partial (3%)
Length = 805
Score = 41.6 bits (96), Expect = 0.002
Identities = 30/128 (23%), Positives = 52/128 (40%)
Frame = +3
Query: 560 PTTAAAPKGKNVAEASIAAATEPTAVPASTPASAGATAAVAAESSVGATAASAGVNATKA 619
P A P G +A + PT+ +S AS+ + + +S A++ + + A
Sbjct: 21 PGVAQTPSGNGIAPSPYQF-NNPTSPSSSPSASSPVSTTTSPPASTPASSPVSTTTSPPA 197
Query: 620 AASSDTPIGEKEKENETPKSPPHQEAPPSPPPTRDAGSMPSPPHQGEKSCPGAATTSEAA 679
+ + +P+ P +PP+P P S SP S P AAT S ++
Sbjct: 198 STPASSPV-------------PTTTSPPAPTPASSPVSTNSPTASPAGSLPAAATPSPSS 338
Query: 680 QIEQAPAP 687
+P+P
Sbjct: 339 TTAGSPSP 362
>TC80366 similar to GP|13877617|gb|AAK43886.1 protein kinase-like protein
{Arabidopsis thaliana}, partial (16%)
Length = 814
Score = 40.8 bits (94), Expect = 0.003
Identities = 28/102 (27%), Positives = 38/102 (36%)
Frame = +3
Query: 603 SSVGATAASAGVNATKAAASSDTPIGEKEKENETPKSPPHQEAPPSPPPTRDAGSMPSPP 662
SS S+ +T A TP TP SPP P +PPP+ + S PSPP
Sbjct: 3 SSPPPATPSSPPPSTPANPPPVTPSAPPPSTPATPSSPPPSTTPSAPPPSTPSNSPPSPP 182
Query: 663 HQGEKSCPGAATTSEAAQIEQAPAPEVGSSSYYNMLPNAIEP 704
S P T+ + P+ + S + P P
Sbjct: 183 TTPAISPPSGGGTTPSPPSRTPPSSDDSPSPPSSKTPPPPSP 308
Score = 34.7 bits (78), Expect = 0.19
Identities = 25/103 (24%), Positives = 40/103 (38%)
Frame = +3
Query: 560 PTTAAAPKGKNVAEASIAAATEPTAVPASTPASAGATAAVAAESSVGATAASAGVNATKA 619
P T ++P A P+A P STPA+ + S+ + S +
Sbjct: 15 PATPSSPPPSTPANPP---PVTPSAPPPSTPATPSSPPPSTTPSAPPPSTPSNSPPSPPT 185
Query: 620 AASSDTPIGEKEKENETPKSPPHQEAPPSPPPTRDAGSMPSPP 662
+ P G + ++PP + PSPP ++ PSPP
Sbjct: 186 TPAISPPSGGGTTPSPPSRTPPSSDDSPSPPSSK-TPPPPSPP 311
>BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xylostella
granulovirus}, partial (42%)
Length = 909
Score = 40.8 bits (94), Expect = 0.003
Identities = 55/222 (24%), Positives = 76/222 (33%), Gaps = 35/222 (15%)
Frame = +3
Query: 504 KARERRMARFSPKFNTEGDAKKRGGAENQAESTKAPKRRRL--TKTSSGAGTSNPGAQPT 561
K R+ + P+ T A G NQ +T PK++ T T + P PT
Sbjct: 99 KKSPRKKEKPPPRPATPPPAPAPPGPPNQTTNTTNPKKQHTPTTPTHPPRPHTPPTPPPT 278
Query: 562 TAAAPKGKNVAEASIAAATEPTA-----VPASTPASAGATAAVAAESSVGATAASAG--V 614
T P + + T PTA PA P G G
Sbjct: 279 TQTTPPREAPTDPGTTRGTPPTADDAQTSPAHHPGEGGGKGGAPPPPPRPPPRHGRGGHQ 458
Query: 615 NATKAAASSDT-PIGEKEKENETPKS----PPHQE---------APPSPPPTRDAGS--- 657
N T+ A + T P G++++ P+ PPHQ+ PPP AG
Sbjct: 459 NQTREAKRTTTEPQGKRDRPRTDPRKKKHPPPHQKEGGNTQKEPRGEDPPPHTPAGGGGG 638
Query: 658 ----MPSPPHQ----GEKSCP-GAATTSEAAQIEQAPAPEVG 690
P PP + GE+ P GA E + +Q A G
Sbjct: 639 PGGPRPPPPKKTARGGEERPPAGARARREKTRGDQQEAAPAG 764
Score = 35.4 bits (80), Expect = 0.11
Identities = 42/161 (26%), Positives = 56/161 (34%), Gaps = 25/161 (15%)
Frame = +1
Query: 528 GAENQAESTKAPKRRRLTKTSSGAGTSN---PGAQPTTAAAPKGKNVAEASIAAATEPTA 584
G + T+ K+R + GT + PG + T K + S T P
Sbjct: 436 GTGGEDTKTRPGKQREPQQNPRERGTGHARTPGKKNTPPHTKKKGETHKKSPGGRTPP-- 609
Query: 585 VPASTPASAGATAAVAA-------ESSVGATAASAGVNATKAAASSDT--PIGEKEK--- 632
P P GA A A E + G K AA+S P G +EK
Sbjct: 610 -PTPRPGGGGARGAPAPPPQKKQREGGKSGPPRAPGHEEKKPAATSRKRPPRGAREKNPA 786
Query: 633 -------ENETPKSPP--HQEAPP-SPPPTRDAGSMPSPPH 663
+ +TPK P H PP +PPP + P PH
Sbjct: 787 AHAREREKKKTPKKNPQNHNNNPPKTPPPQKKHPPPPKTPH 909
Score = 33.1 bits (74), Expect = 0.55
Identities = 20/71 (28%), Positives = 26/71 (36%), Gaps = 3/71 (4%)
Frame = +1
Query: 637 PKSPPHQEAPPSPPPTRDA---GSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSSS 693
P PP +APP+ PPTR + P PPH P P P+
Sbjct: 148 PPPPPPPQAPPTKPPTRQTPKNNTPPPPPHTPPDPTP------------PQPPPQPPKPP 291
Query: 694 YYNMLPNAIEP 704
++ P EP
Sbjct: 292 HHEKRPRTPEP 324
Score = 33.1 bits (74), Expect = 0.55
Identities = 32/142 (22%), Positives = 45/142 (31%)
Frame = +1
Query: 511 ARFSPKFNTEGDAKKRGGAENQAESTKAPKRRRLTKTSSGAGTSNPGAQPTTAAAPKGKN 570
AR K NT KK+G ++ + P T G G A P +G
Sbjct: 520 ARTPGKKNTPPHTKKKGETHKKSPGGRTPPP---TPRPGGGGARGAPAPPPQKKQREG-- 684
Query: 571 VAEASIAAATEPTAVPASTPASAGATAAVAAESSVGATAASAGVNATKAAASSDTPIGEK 630
+ P P AT+ GA + +A + +
Sbjct: 685 -------GKSGPPRAPGHEEKKPAATSRKRPPR--GAREKNPAAHAREREKKKTPKKNPQ 837
Query: 631 EKENETPKSPPHQEAPPSPPPT 652
N PK+PP Q+ P PP T
Sbjct: 838 NHNNNPPKTPPPQKKHPPPPKT 903
Score = 32.0 bits (71), Expect = 1.2
Identities = 38/160 (23%), Positives = 46/160 (28%), Gaps = 23/160 (14%)
Frame = +3
Query: 526 RGGAENQAESTKAPKRRRLTKTSSGAGTSNPGAQPTTAAAPKGKNVAEASIAAATEPTAV 585
RGG +NQ K T T P P P +
Sbjct: 444 RGGHQNQTREAKR------TTTEPQGKRDRPRTDPRKKKHPPPHQKEGGNTQKEPRGEDP 605
Query: 586 PASTPASAGATAAVAAESSVGATAAS------AGVNATKAAASSDT------------PI 627
P TPA G TA AG A + D P
Sbjct: 606 PPHTPAGGGGGPGGPRPPPPKKTARGGEERPPAGARARREKTRGDQQEAAPAGRPRKKPR 785
Query: 628 GEKEKE----NETPKSP-PHQEAPPSPPPTRDAGSMPSPP 662
G +E+E N K P P Q+ P +PPPT+ P P
Sbjct: 786 GAREREGKKKNPQKKPPKPQQQPPKNPPPTKKTPPPPKNP 905
Score = 30.0 bits (66), Expect = 4.7
Identities = 19/48 (39%), Positives = 23/48 (47%), Gaps = 1/48 (2%)
Frame = +3
Query: 618 KAAASSDTPIGEKEKENETPKSPPHQEAPPSPPPT-RDAGSMPSPPHQ 664
K S P G K+ T KSP +E PP P T A + P PP+Q
Sbjct: 48 KGHPSQKPPTGRSHKK--TKKSPRKKEKPPPRPATPPPAPAPPGPPNQ 185
Score = 29.6 bits (65), Expect = 6.1
Identities = 20/62 (32%), Positives = 29/62 (46%), Gaps = 5/62 (8%)
Frame = +1
Query: 635 ETPKSPPHQEAPPSPPPTRDAGSMPSPPHQGEKSCPGAATTSE-----AAQIEQAPAPEV 689
+ PK P H++ P +P P G P PP +++ P TT E A+ PAP
Sbjct: 274 QPPKPPHHEKRPRTPEP---PGGRPPPPTTPKQARP---TTQERGGARGARPRPPPAPPQ 435
Query: 690 GS 691
G+
Sbjct: 436 GT 441
>NP212732 NP212732|AF106929.1|AAD39890.1 putative cell wall protein
Length = 576
Score = 40.4 bits (93), Expect = 0.003
Identities = 32/94 (34%), Positives = 47/94 (49%), Gaps = 9/94 (9%)
Frame = +1
Query: 523 AKKRGGAENQAESTKAPK---RRRLTKTSSGAGTSNPGAQPTTAAAPKGKNVAEASI--- 576
AK + A + A+ APK + +T + A + PGA P TA AP G A+
Sbjct: 226 AKGKDAAPDGAKGKDAPKEGAKGTVTPPAPAAPGAAPGAAPGTAPAPGGPPPEGAAPSPA 405
Query: 577 ---AAATEPTAVPASTPASAGATAAVAAESSVGA 607
AAA P A ++ A AGA+ + AA+++ GA
Sbjct: 406 KGGAAAPTPGAGTGTSVAPAGASGSTAAKTATGA 507
Score = 32.7 bits (73), Expect = 0.72
Identities = 18/64 (28%), Positives = 28/64 (43%), Gaps = 3/64 (4%)
Frame = +1
Query: 619 AAASSDTPIGEKEKENETPKSPPHQEAPPSPPPTRDAGSMPSP---PHQGEKSCPGAATT 675
A ++ P+G P + P APP PP +DA +P +G+ + P A
Sbjct: 55 ACVAAQGPVGAPPAPGTPPPAEPAPGAPPPPPKGKDAKGKDTPDGDAAKGKDAAPDGAKG 234
Query: 676 SEAA 679
+AA
Sbjct: 235 KDAA 246
>TC77143 similar to GP|559557|gb|AAA51649.1|| arabinogalactan-protein {Pyrus
communis}, partial (30%)
Length = 956
Score = 40.0 bits (92), Expect = 0.005
Identities = 35/110 (31%), Positives = 44/110 (39%), Gaps = 3/110 (2%)
Frame = +3
Query: 556 PGAQPTTAAAPKGKNVAEASIAAATEPTAVPASTPASAGATAAVAAESSVGATAASAGVN 615
PGA PT + A T PT P +TP +A T A + A+A
Sbjct: 180 PGAAPTQPPS-----------ATPTPPTPAPVATPPTA--TPPTATPPTATPPPAAAPTP 320
Query: 616 ATKAAASSD---TPIGEKEKENETPKSPPHQEAPPSPPPTRDAGSMPSPP 662
AT A A+S TP + + T SPP AP P GS +PP
Sbjct: 321 ATPAPATSPPAPTPTSDAPTPDSTSSSPP---APGPGGPAPGPGSTDAPP 461
Score = 35.4 bits (80), Expect = 0.11
Identities = 37/155 (23%), Positives = 56/155 (35%)
Frame = +3
Query: 572 AEASIAAATEPTAVPASTPASAGATAAVAAESSVGATAASAGVNATKAAASSDTPIGEKE 631
A+A AA T+P P++TP ++ TA AAA
Sbjct: 171 AQAPGAAPTQP---PSATPTPPTPAPVATPPTATPPTATPPTATPPPAAAP--------- 314
Query: 632 KENETPKSPPHQEAPPSPPPTRDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGS 691
TP +P +PP+P PT DA + P + ++S A PAP GS
Sbjct: 315 ----TPATPAPATSPPAPTPTSDAPT------------PDSTSSSPPAPGPGGPAPGPGS 446
Query: 692 SSYYNMLPNAIEPSEFLLAGLNRDVIEKEVLSRGL 726
+ PS +N+ ++ LS +
Sbjct: 447 TD--------APPSPSAAFSINKPIMAATALSAAI 527
>BQ141681 weakly similar to GP|15145793|gb| basic proline-rich protein {Sus
scrofa}, partial (9%)
Length = 516
Score = 39.3 bits (90), Expect = 0.008
Identities = 32/107 (29%), Positives = 38/107 (34%)
Frame = +2
Query: 556 PGAQPTTAAAPKGKNVAEASIAAATEPTAVPASTPASAGATAAVAAESSVGATAASAGVN 615
PGA PT K KN P +T TA E AA AG
Sbjct: 134 PGANPT-----KSKNTDPKPEGPTKGPNQRAKTTHKR---TARGKGEEPPEGEAARAGDE 289
Query: 616 ATKAAASSDTPIGEKEKENETPKSPPHQEAPPSPPPTRDAGSMPSPP 662
+ A P G + E K PP + PP+ PP D P+PP
Sbjct: 290 ENREKAGPQPPRGPRRPE----KDPPPRAEPPAAPPPADPEGRPAPP 418
Score = 30.4 bits (67), Expect = 3.6
Identities = 22/84 (26%), Positives = 29/84 (34%)
Frame = +3
Query: 587 ASTPASAGATAAVAAESSVGATAASAGVNATKAAASSDTPIGEKEKENETPKSPPHQEAP 646
A T T AE A AT+ P ++ K+PP +P
Sbjct: 195 AQTKGQKQPTKERPAEKEKNRQKAKQRARATRKTGRKRDPNPPGDR-GAPKKTPPPGPSP 371
Query: 647 PSPPPTRDAGSMPSPPHQGEKSCP 670
P PP R + P PP +G P
Sbjct: 372 PPRPPRRTRRAGPPPPGRGAAPPP 443
>TC85251 homologue to PIR|JQ2128|JQ2128 metallothionein - soybean, partial
(93%)
Length = 1554
Score = 39.3 bits (90), Expect = 0.008
Identities = 41/169 (24%), Positives = 55/169 (32%), Gaps = 28/169 (16%)
Frame = -2
Query: 547 TSSGAGTSNPGAQPTTAAAPKGKNVAEASIAAATEPTAVPASTP---------------- 590
TS T P + P T+ APK + + P + P STP
Sbjct: 752 TSQPPVTVAPKSAPVTSPAPKIAPASSPKVPPPQPPKSSPVSTPTLPPPLPPPPKISPTP 573
Query: 591 -----------ASAGATAAVAAESSVGATAASAGVNATKAAASSDTPIGEKEKENETPKS 639
A+ A A ++ A A S + A A + P E K
Sbjct: 572 VQTPPAPAPVKATPVPAPAPAKQAPTPAPATSPPIPAPTPAIEAPVPAPESSKPKRRRHR 393
Query: 640 PPHQE-APPSPPPTRDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAP 687
P H+ P+P PT S P+PP T A + APAP
Sbjct: 392 PKHRRHQAPAPAPTVIHKSPPAPP------------TDTTADSDTAPAP 282
Score = 37.0 bits (84), Expect = 0.038
Identities = 36/118 (30%), Positives = 52/118 (43%), Gaps = 5/118 (4%)
Frame = -2
Query: 580 TEPTAV-PAST-PASAGATAAVAAESSVGATAASAGVN--ATKAAASSDTPIGEKEKENE 635
T PTA+ P +T P + A+ + ++ V SA V A K A +S + +
Sbjct: 818 TTPTAITPVTTQPPTVVASPPITSQPPVTVAPKSAPVTSPAPKIAPASSPKVPPPQPPKS 639
Query: 636 TPKSPPHQEAPPSPPPTRDAGSMPSPPHQGEKSCPGAATTSEA-AQIEQAPAPEVGSS 692
+P S P PP PPP + + P+P P AT A A +QAP P +S
Sbjct: 638 SPVSTP-TLPPPLPPPPKIS---PTPVQTPPAPAPVKATPVPAPAPAKQAPTPAPATS 477
>TC76720 similar to GP|11121502|emb|CAC14888. putative extensin {Nicotiana
sylvestris}, partial (51%)
Length = 1290
Score = 38.9 bits (89), Expect = 0.010
Identities = 27/98 (27%), Positives = 40/98 (40%), Gaps = 2/98 (2%)
Frame = +1
Query: 598 AVAAESSVGATAASAGVNA-TKAAASSDTPIGEKEKENETPKSPPHQEAPPSPPPTRDAG 656
A +A+S A ++ +N+ + + +P TP PH ++PP+P P
Sbjct: 544 ASSADSPAPTPATNSSLNSPSPTPIPTPSPANSPPAPTPTPTPSPHSDSPPAPSPDNSPS 723
Query: 657 SMPSP-PHQGEKSCPGAATTSEAAQIEQAPAPEVGSSS 693
S PSP P P A + A I E G SS
Sbjct: 724 SSPSPSPSSSPAPSPDEAADNNA--ISHTGIGEDGKSS 831
>TC80891 similar to GP|21622328|emb|CAD36969. probable isoamyl alcohol
oxidase {Neurospora crassa}, partial (12%)
Length = 766
Score = 38.9 bits (89), Expect = 0.010
Identities = 22/59 (37%), Positives = 27/59 (45%), Gaps = 2/59 (3%)
Frame = +1
Query: 636 TPKSPPHQEAPPSPPPTRDAGSMPSPPHQGEKSCPGAATTSEAA--QIEQAPAPEVGSS 692
TP PP PPS PP+ S PSP + P A T ++A + PAP SS
Sbjct: 358 TPAGPPRPSGPPSTPPSAAVSSPPSPSAPSATTAPLAPLTRQSAPRSRPRGPAPRRTSS 534
>TC89270 similar to GP|22773423|gb|AAL66352.2 catalase-peroxidase
{Neurospora crassa}, partial (25%)
Length = 792
Score = 38.1 bits (87), Expect = 0.017
Identities = 44/174 (25%), Positives = 57/174 (32%), Gaps = 9/174 (5%)
Frame = +1
Query: 535 STKAPKRRRLTKTSSGAGTSNPG-AQPTTAAAPKGKNVAEASIA-AATEPTAVPASTPAS 592
+T P R T+ S TS P P T P A S A T P +T A
Sbjct: 259 TTSPPSSRSTTRASRR--TSRP**PTPRTGGLPTLATTAVCSSAWPGTAPARTEFTTDAE 432
Query: 593 AGATAAVAAESSVGATAASAGVNATKAAASSDTPIGEKEKENET-------PKSPPHQEA 645
A+ A+ S SA S + + + P SP
Sbjct: 433 VVERASNASHRSTAGRTMSASTRPVGCCGPSSKSTATRSRGPTS*SWPATWPSSPWVSRR 612
Query: 646 PPSPPPTRDAGSMPSPPHQGEKSCPGAATTSEAAQIEQAPAPEVGSSSYYNMLP 699
P SP G P+ P G PG+ATTS +A A S++ P
Sbjct: 613 PASPEAVPTPGK-PTSPSTGAARTPGSATTSATPTATRARADRASSTARRRQSP 771
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.313 0.129 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,807,845
Number of Sequences: 36976
Number of extensions: 506150
Number of successful extensions: 7461
Number of sequences better than 10.0: 244
Number of HSP's better than 10.0 without gapping: 4374
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6227
length of query: 1016
length of database: 9,014,727
effective HSP length: 106
effective length of query: 910
effective length of database: 5,095,271
effective search space: 4636696610
effective search space used: 4636696610
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0029b.2


