

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
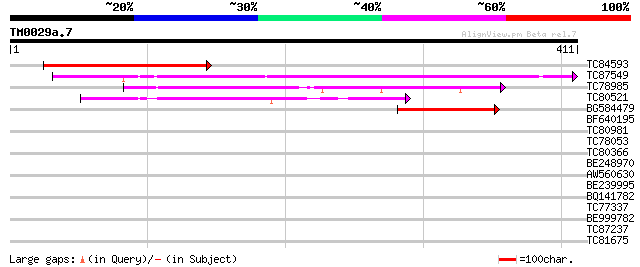
Query= TM0029a.7
(411 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC84593 similar to GP|15810016|gb|AAL06935.1 AT3g58520/F14P22_11... 209 2e-54
TC87549 similar to PIR|T01734|T01734 hypothetical protein A_IG00... 146 2e-35
TC78985 similar to PIR|T06001|T06001 hypothetical protein F17M5.... 129 3e-30
TC80521 weakly similar to GP|18377712|gb|AAL67006.1 unknown prot... 86 3e-17
BG584479 similar to GP|9758299|dbj| gene_id:MJH22.4~pir||T01734~... 45 4e-05
BF640195 similar to GP|13374856|em putative protein {Arabidopsis... 37 0.011
TC80981 similar to GP|8698730|gb|AAF78488.1| ESTs gb|Z18526 gb|... 34 0.091
TC78053 similar to GP|5923675|gb|AAD56326.1| putative mRNA cappi... 31 0.77
TC80366 similar to GP|13877617|gb|AAK43886.1 protein kinase-like... 30 1.7
BE248970 30 2.2
AW560630 weakly similar to GP|21593023|gb| unknown {Arabidopsis ... 29 3.8
BE239995 weakly similar to GP|4008010|gb|A receptor-like protein... 28 5.0
BQ141782 similar to GP|4028979|emb| glutamate permease {syntheti... 28 6.5
TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Atel... 28 6.5
BE999782 similar to PIR|T09661|T096 ascorbate oxidase promoter-b... 28 8.5
TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-inf... 28 8.5
TC81675 similar to GP|4586254|emb|CAB40995.1 auxilin-like protei... 28 8.5
>TC84593 similar to GP|15810016|gb|AAL06935.1 AT3g58520/F14P22_110
{Arabidopsis thaliana}, partial (29%)
Length = 457
Score = 209 bits (531), Expect = 2e-54
Identities = 103/122 (84%), Positives = 108/122 (88%)
Frame = +3
Query: 25 KVRLKWVKNRSLDHIIDKETDLKAASLLKDAINRSSTGFLTAKSFSGWQKLLGLTVPVLR 84
K LKWVKNRS+DHIIDKETDLKAASLLKDAI RSST FLTAK+FS WQKLLGLTVPVLR
Sbjct: 90 KYXLKWVKNRSIDHIIDKETDLKAASLLKDAIKRSSTSFLTAKTFSDWQKLLGLTVPVLR 269
Query: 85 FIRRYPTLFHEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNHHENDTVERLSKFLMMTKT 144
FIRRYPTLFHEFPHPRW SLPCFRLTDTA LL SQEQS++ HEND VER SK LMM K+
Sbjct: 270 FIRRYPTLFHEFPHPRWNSLPCFRLTDTAQLLDSQEQSIYAVHENDIVERFSKVLMMMKS 449
Query: 145 RT 146
RT
Sbjct: 450 RT 455
>TC87549 similar to PIR|T01734|T01734 hypothetical protein A_IG002N01.30 -
Arabidopsis thaliana (fragment), partial (38%)
Length = 1766
Score = 146 bits (368), Expect = 2e-35
Identities = 109/383 (28%), Positives = 188/383 (48%), Gaps = 3/383 (0%)
Frame = +2
Query: 32 KNRSLDHIIDKETDLKAASLLKDAINRSSTGFLTAKSFSGWQKLLGLTVP--VLRFIRRY 89
K D++I K+ LK ++ + T ++ + +++ LGL ++ +RR+
Sbjct: 467 KELPFDNVIQKDKKLKFVLKVRKILVSKPTRVMSLQELGKYRRELGLDKKRKLIVILRRF 646
Query: 90 PTLFHEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNHHENDTVERLSKFLMMTKTRTVPL 149
P +F E SL F++T A L+ +E + N E+ V +L K LMM + + L
Sbjct: 647 PGVF-EIVEDGCYSLK-FKMTSEAEKLYLEELRVRNEMEDVVVTKLRKLLMMPLEKRILL 820
Query: 150 VSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPNGVSAVRLSKWCEEFAVSALQKSNER 209
+ L D+GLP F T+ ++P+ F+ V+ G A+ L+ W AVSA + S E
Sbjct: 821 EKIGHLANDLGLPREFRDTICHRYPEFFKVVQTERG-PALELTHWDPHLAVSAAELSAEE 997
Query: 210 DG-GDDGYRDFKRGKAALVFPMRFPRGYGAQKKVRFWMDEFHKLPYISPYADSSKIDPNS 268
+ + ++ +A ++ P+G K + +F +PYISPY+D S I N+
Sbjct: 998 NRIREVEEQNLIIDRAPKFNRVKLPKGLSLSKGEMRKIMQFRDIPYISPYSDFSMIGLNT 1177
Query: 269 DLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRIFTRYPGIFYLSLKCKT 328
EK GV+HE+LSLT+ K+T + REE + + R+P +FY+SLK
Sbjct: 1178PEKEKHACGVIHEILSLTLEKRTLVDNFTHFREEFRFSQQLRGMLIRHPDMFYISLKGDR 1357
Query: 329 TTVTLREGYARGKLVDPHPLVRHRDKFYHVMKTGLLYRGDGSIPKPEGGALLLDEIGDEG 388
+V LRE Y +LVD L+ ++K ++ +G G+ +G + ++I E
Sbjct: 1358DSVFLREAYRDSQLVDKDRLLLVKEKLRSLVDIPRFPKGRGAGRTXDGDGMGENDI--EN 1531
Query: 389 SEEEEVETSDEFSEGEVSGSDSE 411
++E E E+S+ + SD +
Sbjct: 1532RQDESGEEEQEWSDADDLISDDD 1600
>TC78985 similar to PIR|T06001|T06001 hypothetical protein F17M5.260 -
Arabidopsis thaliana, partial (33%)
Length = 2496
Score = 129 bits (323), Expect = 3e-30
Identities = 98/289 (33%), Positives = 145/289 (49%), Gaps = 12/289 (4%)
Frame = +1
Query: 83 LRFIRRYPTLFHEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNHHENDTVERLSKFLMMT 142
+ FI ++P +F F HP L C RLT A+L QE+ V RL K +MM+
Sbjct: 406 IAFILKFPHVFEIFEHPVQRILFC-RLTRKAILQIEQERLALADQVERAVTRLRKMIMMS 582
Query: 143 KTRTVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPNGVSA-VRLSKWCEEFAVS 201
+ L + + +GLPD FE +++ ++P+ F+ V A + + L ++ + A
Sbjct: 583 NGLRLRLEHVRIARSALGLPDDFEYSVVLRYPEFFRLVDAKETRNKYIELVEFDPKLAKC 762
Query: 202 ALQKSNERDGGDDGYRDFKRGKAA----LVFPMRFPRGYGAQKKVRFWMDEFHKLPYISP 257
A++ + ER YR+ RG A F + FP G+ K R M ++ +LPY SP
Sbjct: 763 AIEDARERV-----YRE--RGSEAEDIRFSFLIDFPPGFKISKYFRIAMWKWQRLPYWSP 921
Query: 258 YADSSKIDPNS----DLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRIF 313
Y D D S MEKR V +HELLSLT+ KK + R +NLP K I
Sbjct: 922 YEDVLGYDLRSIEAQKRMEKRAVATIHELLSLTVEKKITLERIAHFRMAMNLPKKLKEIL 1101
Query: 314 TRYPGIFYLSLK---CKTTTVTLREGYARGKLVDPHPLVRHRDKFYHVM 359
++ GIFY+S + K TV LRE Y +G+LV+P+ L R K ++
Sbjct: 1102LQHQGIFYVSTRGNHGKLHTVFLREAYRKGELVEPNDLYLARRKLVELV 1248
>TC80521 weakly similar to GP|18377712|gb|AAL67006.1 unknown protein
{Arabidopsis thaliana}, partial (57%)
Length = 1025
Score = 85.5 bits (210), Expect = 3e-17
Identities = 68/241 (28%), Positives = 113/241 (46%), Gaps = 2/241 (0%)
Frame = +2
Query: 52 LKDAINRSSTGFLTAKSFSGWQKLLGLTVPVLRFIRRYPTLFHEFPHPRWPSLPCFRLTD 111
+K+ I R + ++ K L L L +I +YP+ F +F LT
Sbjct: 356 IKNIILRYPNNQIPIQTLQKKFKTLDLQGKALNWISKYPSCF-QFHQDH------VLLTK 514
Query: 112 TALLLHSQEQSLHNHHENDTVERLSKFLMMTKTRTVPLVSLYPLKWDIGLPDSFEKTLIP 171
+ L +EQSL + E+ V RL+K LM++ + ++ + +K +G PD + ++
Sbjct: 515 RMMELVHEEQSLKDSLESVFVPRLAKLLMLSLNNCLNVMKINEIKNSLGFPDDYLIGIVA 694
Query: 172 KFPDQFQFVKAPNGVSA--VRLSKWCEEFAVSALQKSNERDGGDDGYRDFKRGKAALVFP 229
K+PD F+ S+ V L KW +FAVS ++ ++G + + + P
Sbjct: 695 KYPDLFRIRNESGRRSSMVVELMKWNPDFAVSEVEALAMKNGVEVNF--------SCCLP 850
Query: 230 MRFPRGYGAQKKVRFWMDEFHKLPYISPYADSSKIDPNSDLMEKRVVGVLHELLSLTMHK 289
+ + EF +PY+SPY+D + S MEKR VG++HELL LT+ K
Sbjct: 851 SSWVKSLEK-------FHEFELVPYVSPYSDPRGLVEGSKEMEKRNVGLVHELLXLTLWK 1009
Query: 290 K 290
K
Sbjct: 1010K 1012
>BG584479 similar to GP|9758299|dbj| gene_id:MJH22.4~pir||T01734~similar to
unknown protein {Arabidopsis thaliana}, partial (16%)
Length = 786
Score = 45.4 bits (106), Expect = 4e-05
Identities = 24/75 (32%), Positives = 45/75 (60%), Gaps = 1/75 (1%)
Frame = +1
Query: 282 LLSLTMHKKTKRNYLRSLREELNLPHKFTRIFTRYPGIFYLSLKCKTTTVTLREGY-ARG 340
+L +T+ K+T ++L R+E LP+K + R+P +FY SLK + +V L E + +G
Sbjct: 1 VLGMTIEKRTLIDHLTHFRKEFGLPNKLRGMIVRHPELFYTSLKGQRDSVFLVERFDEKG 180
Query: 341 KLVDPHPLVRHRDKF 355
L++ ++ +DK+
Sbjct: 181 NLLEKDEVLALQDKW 225
>BF640195 similar to GP|13374856|em putative protein {Arabidopsis thaliana},
partial (32%)
Length = 521
Score = 37.4 bits (85), Expect = 0.011
Identities = 30/104 (28%), Positives = 51/104 (48%), Gaps = 2/104 (1%)
Frame = +2
Query: 64 LTAKSFSGWQKLLGLTVP--VLRFIRRYPTLFHEFPHPRWPSLPCFRLTDTALLLHSQEQ 121
+ +S S ++K + L P + F+R+ P LF E R L C LT A +L + +
Sbjct: 200 IPVRSLSHYRKQISLPKPHRISDFLRKTPKLF-ELYKDRNGVLWC-GLTQKAEVLMEEHK 373
Query: 122 SLHNHHENDTVERLSKFLMMTKTRTVPLVSLYPLKWDIGLPDSF 165
+ +E+ E +++ LMM+ + V L + + D GLP F
Sbjct: 374 RVIEENEDKAAEYVTRLLMMSVDKRVQLDKIAHFRRDFGLPMDF 505
>TC80981 similar to GP|8698730|gb|AAF78488.1| ESTs gb|Z18526 gb|AI994480
gb|T44186 gb|Z30806 come from this gene. {Arabidopsis
thaliana}, partial (37%)
Length = 699
Score = 34.3 bits (77), Expect = 0.091
Identities = 14/46 (30%), Positives = 29/46 (62%)
Frame = +3
Query: 366 RGDGSIPKPEGGALLLDEIGDEGSEEEEVETSDEFSEGEVSGSDSE 411
+G G + +G +++E ++ + ++ + SD+ S+G VSGSDS+
Sbjct: 315 KGKGIMRDDKGKGKMIEEEDEDDDDSDDSDDSDDDSDGNVSGSDSD 452
>TC78053 similar to GP|5923675|gb|AAD56326.1| putative mRNA capping enzyme
RNA guanylyltransferase {Arabidopsis thaliana}, partial
(71%)
Length = 2017
Score = 31.2 bits (69), Expect = 0.77
Identities = 19/45 (42%), Positives = 27/45 (59%)
Frame = -1
Query: 153 YPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPNGVSAVRLSKWCEE 197
YPL ++ L DS E+ L + PDQ F+K VS V +S +C+E
Sbjct: 718 YPLFLNLSL-DSIERYLSNQSPDQVFFLKLSVDVSLV*MSIFCQE 587
>TC80366 similar to GP|13877617|gb|AAK43886.1 protein kinase-like protein
{Arabidopsis thaliana}, partial (16%)
Length = 814
Score = 30.0 bits (66), Expect = 1.7
Identities = 18/50 (36%), Positives = 26/50 (52%)
Frame = -1
Query: 362 GLLYRGDGSIPKPEGGALLLDEIGDEGSEEEEVETSDEFSEGEVSGSDSE 411
G+ GDG +P P+GG + +G EG E E V+ ++G V G E
Sbjct: 247 GVREGGDGVVPPPDGGD-IAGVVGGEGGEFEGVDGGG--ADGVVDGGGDE 107
>BE248970
Length = 515
Score = 29.6 bits (65), Expect = 2.2
Identities = 14/34 (41%), Positives = 20/34 (58%)
Frame = -2
Query: 94 HEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNHH 127
H+ P+P+ LP F T LLHS E+ +H+ H
Sbjct: 460 HQLPNPKESLLP-FIGVPTIFLLHSSEKFIHHQH 362
>AW560630 weakly similar to GP|21593023|gb| unknown {Arabidopsis thaliana},
partial (19%)
Length = 525
Score = 28.9 bits (63), Expect = 3.8
Identities = 17/53 (32%), Positives = 28/53 (52%)
Frame = -3
Query: 349 VRHRDKFYHVMKTGLLYRGDGSIPKPEGGALLLDEIGDEGSEEEEVETSDEFS 401
++ RDK+ H GLL+ S+ K + +G ++EE+VE+S FS
Sbjct: 328 IKRRDKWIHPKA*GLLHTSKRSV*KHRE-----ERVGCSKTQEEQVESSSLFS 185
>BE239995 weakly similar to GP|4008010|gb|A receptor-like protein kinase
{Arabidopsis thaliana}, partial (4%)
Length = 453
Score = 28.5 bits (62), Expect = 5.0
Identities = 15/63 (23%), Positives = 29/63 (45%)
Frame = +3
Query: 348 LVRHRDKFYHVMKTGLLYRGDGSIPKPEGGALLLDEIGDEGSEEEEVETSDEFSEGEVSG 407
L H D ++ ++ + +PKP+ LL+ ++ EG +TS +E +S
Sbjct: 87 LQNHADDRPNMTSVVVMLSSENGLPKPKEPGLLIKKLSTEGEASSGRQTSSSTNEVTISL 266
Query: 408 SDS 410
D+
Sbjct: 267 LDA 275
>BQ141782 similar to GP|4028979|emb| glutamate permease {synthetic
construct}, partial (4%)
Length = 1259
Score = 28.1 bits (61), Expect = 6.5
Identities = 17/46 (36%), Positives = 23/46 (49%), Gaps = 5/46 (10%)
Frame = -2
Query: 71 GWQKLLGLTVPVLRFIRRY-----PTLFHEFPHPRWPSLPCFRLTD 111
G K LG + ++ +R+Y P L E P PRW S CF L +
Sbjct: 211 GLFKRLGFRLFIVPRVRKYSCSRAPPLVIE*PPPRWSSSFCFPLVE 74
>TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Ateline
herpesvirus 3}, partial (8%)
Length = 986
Score = 28.1 bits (61), Expect = 6.5
Identities = 15/43 (34%), Positives = 23/43 (52%), Gaps = 5/43 (11%)
Frame = +3
Query: 374 PEGGALLLDEIGDE-----GSEEEEVETSDEFSEGEVSGSDSE 411
PEGGA DE G+E G +++E + DE + E G + +
Sbjct: 558 PEGGAGGADENGEEEDDEDGDDQDEDDDEDEDDDDEEEGGEED 686
>BE999782 similar to PIR|T09661|T096 ascorbate oxidase promoter-binding
protein AOBP - winter squash, partial (17%)
Length = 630
Score = 27.7 bits (60), Expect = 8.5
Identities = 18/81 (22%), Positives = 33/81 (40%)
Frame = +2
Query: 318 GIFYLSLKCKTTTVTLREGYARGKLVDPHPLVRHRDKFYHVMKTGLLYRGDGSIPKPEGG 377
G+ + SL C T T R PL + ++ G+ +PE
Sbjct: 251 GVHHPSLNCNGTVFTFRTDT---------PLCESMESALNLADQGVNISQKNGFIRPEAL 403
Query: 378 ALLLDEIGDEGSEEEEVETSD 398
+ + +G+E S+E +++SD
Sbjct: 404 RIHVPYVGEEKSDEHSIKSSD 466
>TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-infected
erythrocyte surface antigen {Plasmodium falciparum},
partial (2%)
Length = 2007
Score = 27.7 bits (60), Expect = 8.5
Identities = 17/50 (34%), Positives = 26/50 (52%), Gaps = 6/50 (12%)
Frame = +1
Query: 368 DGSIPKPEGGALLLDEIGDEGSEEEEVETSD------EFSEGEVSGSDSE 411
+ + K EGG EI +E S++E ETS+ E E +GSD++
Sbjct: 283 ENELKKNEGGEKETGEITEEKSKQENEETSETNSKDKENEESNQNGSDAK 432
>TC81675 similar to GP|4586254|emb|CAB40995.1 auxilin-like protein
{Arabidopsis thaliana}, partial (7%)
Length = 848
Score = 27.7 bits (60), Expect = 8.5
Identities = 16/55 (29%), Positives = 25/55 (45%), Gaps = 1/55 (1%)
Frame = -3
Query: 99 PRWPSLPCFRLTDTALLLHSQEQSLHN-HHENDTVERLSKFLMMTKTRTVPLVSL 152
P WP PC LT H +E H+ HH D + + + T+ +P++ L
Sbjct: 660 PSWPQQPCSSLT------HHREDHFHSQHHYCDCCLQQNHQVHQTEMLEMPMLQL 514
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.138 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,785,212
Number of Sequences: 36976
Number of extensions: 214127
Number of successful extensions: 1199
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 1182
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1194
length of query: 411
length of database: 9,014,727
effective HSP length: 98
effective length of query: 313
effective length of database: 5,391,079
effective search space: 1687407727
effective search space used: 1687407727
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0029a.7


