

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0029a.6
(183 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
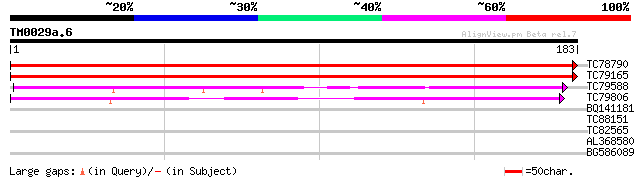
TC78790 similar to PIR|T45678|T45678 hypothetical protein F14P22... 357 1e-99
TC79165 similar to PIR|T45678|T45678 hypothetical protein F14P22... 341 1e-94
TC79588 weakly similar to GP|11994114|dbj|BAB01117. gene_id:MYF2... 74 2e-14
TC79806 similar to GP|12597833|gb|AAG60143.1 hypothetical protei... 68 2e-12
BQ141181 similar to GP|8777430|dbj| dbj|BAA91947.1~gene_id:MHM17... 28 2.0
TC88151 similar to GP|19225025|gb|AAL86501.1 putative endonuclea... 27 4.5
TC82565 similar to GP|6041842|gb|AAF02151.1| unknown protein {Ar... 26 7.7
AL368580 similar to GP|14194151|gb| AT5g40780/K1B16_3 {Arabidops... 26 7.7
BG586089 similar to GP|19225025|gb putative endonuclease {Oryza ... 26 7.7
>TC78790 similar to PIR|T45678|T45678 hypothetical protein F14P22.170 -
Arabidopsis thaliana, partial (69%)
Length = 1681
Score = 357 bits (916), Expect = 1e-99
Identities = 172/183 (93%), Positives = 180/183 (97%)
Frame = +1
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNSNPGSAPHALLAMGKV 60
ANTHVNVHQDLKDVKLWQVHTLLKGLEKIA SADIPMLVCGDFNS PGSAPHALLAMGKV
Sbjct: 808 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAVSADIPMLVCGDFNSVPGSAPHALLAMGKV 987
Query: 61 DPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFARTIGLGYEQHKRRLDSATNEPLFTN 120
DPSHPDLA+DPL+ILRPHSKL+HQLPLVSAYSSFART+GLGYEQHKRR+DS+TNEPLFTN
Sbjct: 988 DPSHPDLAVDPLNILRPHSKLIHQLPLVSAYSSFARTVGLGYEQHKRRMDSSTNEPLFTN 1167
Query: 121 VTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRCCKN 180
VTRDFIG+LDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRCCKN
Sbjct: 1168VTRDFIGSLDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRCCKN 1347
Query: 181 RSR 183
+ R
Sbjct: 1348KPR 1356
>TC79165 similar to PIR|T45678|T45678 hypothetical protein F14P22.170 -
Arabidopsis thaliana, partial (60%)
Length = 1396
Score = 341 bits (874), Expect = 1e-94
Identities = 166/183 (90%), Positives = 173/183 (93%)
Frame = +2
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNSNPGSAPHALLAMGKV 60
ANTHV+V QDLKDVKLWQVHTLLKGLEKIA SADIPMLVCGDFNS PGSAPHALLAMGKV
Sbjct: 590 ANTHVHVQQDLKDVKLWQVHTLLKGLEKIATSADIPMLVCGDFNSVPGSAPHALLAMGKV 769
Query: 61 DPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFARTIGLGYEQHKRRLDSATNEPLFTN 120
DPSHPDL +DPL+ILRPHSKLVHQLPLVSAYSSFART GL +EQHKRRLD TNEPLFT+
Sbjct: 770 DPSHPDLTVDPLNILRPHSKLVHQLPLVSAYSSFARTAGLAFEQHKRRLDGGTNEPLFTS 949
Query: 121 VTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRCCKN 180
VTRDF+G LDYIFYTADSLVVESLLELLDEESLRKDTALPSP WSSDHIALLAEFRCCKN
Sbjct: 950 VTRDFVGTLDYIFYTADSLVVESLLELLDEESLRKDTALPSPGWSSDHIALLAEFRCCKN 1129
Query: 181 RSR 183
+SR
Sbjct: 1130KSR 1138
>TC79588 weakly similar to GP|11994114|dbj|BAB01117.
gene_id:MYF24.23~unknown protein {Arabidopsis thaliana},
partial (40%)
Length = 1742
Score = 74.3 bits (181), Expect = 2e-14
Identities = 62/215 (28%), Positives = 97/215 (44%), Gaps = 36/215 (16%)
Frame = +3
Query: 2 NTHVNVHQDLKDVKLWQVHTLLKGLEKIAAS-ADIPMLVCGDFNSNPGSAPHALLAMGKV 60
N HV + + D+KL QV L+ K++ DIP+++ G+ NS P SA + L+ K+
Sbjct: 696 NIHVLFNPNRGDIKLGQVRLLIDKAYKLSQEWGDIPVILAGELNSVPQSAIYKFLSSSKL 875
Query: 61 D-PSHPDLAIDPLSILRPHSK----------------------------------LVHQL 85
D SH + +RP+ + L HQL
Sbjct: 876 DIQSHDRRNMSGQLEIRPNHREFRSNISMSVSRQLLHKWSAEELILATGAKGVTCLQHQL 1055
Query: 86 PLVSAYSSFARTIGLGYEQHKRRLDSATNEPLFTNVTRDFIGALDYIFYTADSLVVESLL 145
L SAYS +H+ R D EPL T+ F+G +DYI++ ++ L +L
Sbjct: 1056KLRSAYSGVP-------GKHRTRDD--IGEPLATSYHSKFMGTVDYIWH-SEELTPVRVL 1205
Query: 146 ELLDEESLRKDTALPSPEWSSDHIALLAEFRCCKN 180
E L + LR+ LP+ +W SDH+A++ E K+
Sbjct: 1206ETLPVDVLRRTRGLPTEKWGSDHLAVVCELAFAKS 1310
>TC79806 similar to GP|12597833|gb|AAG60143.1 hypothetical protein
{Arabidopsis thaliana}, partial (76%)
Length = 1598
Score = 67.8 bits (164), Expect = 2e-12
Identities = 60/186 (32%), Positives = 76/186 (40%), Gaps = 7/186 (3%)
Frame = +1
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAA------SADIPMLVCGDFNSNPGSAPHAL 54
ANTH+ + DVK+ QV LL L + ++V GDFNS PG +
Sbjct: 727 ANTHIYWDPEWADVKIAQVKYLLSRLSQFKTLVSDRYECKPEVIVAGDFNSQPGDPVYRY 906
Query: 55 LAMGKVDPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFARTIGLGYEQHKRRLDSATN 114
L G +P S L H +PL S Y+S
Sbjct: 907 LISG-----------NPSSELITDCIEEHPIPLSSVYAS------------------TRG 999
Query: 115 EPLFTNVTRDFIGALDYI-FYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLA 173
EP FTN T F G LDYI F +D + S LEL D E+ LP+ SDH+ + A
Sbjct: 1000EPPFTNYTPGFTGTLDYILFCPSDHMKPISYLELPDSEAADIVGGLPNLSHPSDHLPIGA 1179
Query: 174 EFRCCK 179
EF K
Sbjct: 1180EFEIIK 1197
>BQ141181 similar to GP|8777430|dbj| dbj|BAA91947.1~gene_id:MHM17.1~similar
to unknown protein {Arabidopsis thaliana}, partial
(13%)
Length = 585
Score = 28.1 bits (61), Expect = 2.0
Identities = 20/62 (32%), Positives = 29/62 (46%), Gaps = 14/62 (22%)
Frame = +1
Query: 23 LKGLEKIAASADIP-MLVCGDFNS------------NPGSAPH-ALLAMGKVDPSHPDLA 68
++G K++ S P +L+CGD N N + P ALL +G+ P PDL
Sbjct: 1 IRGQSKVSKSVMAPRILLCGDVNGRLNQLFKRVSSVNKSAGPFDALLCVGQFFPDSPDLL 180
Query: 69 ID 70
D
Sbjct: 181DD 186
>TC88151 similar to GP|19225025|gb|AAL86501.1 putative endonuclease {Oryza
sativa (japonica cultivar-group)}, partial (38%)
Length = 889
Score = 26.9 bits (58), Expect = 4.5
Identities = 17/50 (34%), Positives = 29/50 (58%), Gaps = 5/50 (10%)
Frame = +1
Query: 2 NTHVNVHQD--LKDVKLWQVHTLLKGLEKIAAS---ADIPMLVCGDFNSN 46
NTH+ D L V+L QV+ +LK +E+ +P+++CGD+N +
Sbjct: 703 NTHLLFPHDSSLCIVRLDQVYQILKYVEQYQKENRLKPMPIILCGDWNGS 852
>TC82565 similar to GP|6041842|gb|AAF02151.1| unknown protein {Arabidopsis
thaliana}, partial (65%)
Length = 1141
Score = 26.2 bits (56), Expect = 7.7
Identities = 16/57 (28%), Positives = 28/57 (49%)
Frame = +2
Query: 118 FTNVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAE 174
F + RDFIG ++ + SLL+++ + SL T + P W++ +L E
Sbjct: 371 FEKIGRDFIGTRVFLTSLLGIMRTMSLLKMMMKISLL--TGIEDPRWNTTRT*ILLE 535
>AL368580 similar to GP|14194151|gb| AT5g40780/K1B16_3 {Arabidopsis
thaliana}, partial (17%)
Length = 515
Score = 26.2 bits (56), Expect = 7.7
Identities = 9/22 (40%), Positives = 16/22 (71%)
Frame = +1
Query: 11 LKDVKLWQVHTLLKGLEKIAAS 32
+ ++ LW +H +L G EK+AA+
Sbjct: 325 ISNICLWNIHNMLIGGEKVAAT 390
>BG586089 similar to GP|19225025|gb putative endonuclease {Oryza sativa
(japonica cultivar-group)}, partial (47%)
Length = 819
Score = 26.2 bits (56), Expect = 7.7
Identities = 16/50 (32%), Positives = 28/50 (56%), Gaps = 5/50 (10%)
Frame = +1
Query: 2 NTHVNVHQD--LKDVKLWQVHTLLKGLEKIAASADI---PMLVCGDFNSN 46
NTH+ D L V+L QV+ +L+ +E + P+++CGD+N +
Sbjct: 511 NTHLLFPHDATLSLVRLKQVYKILQYVESYQNDFQLKPMPIMLCGDWNGS 660
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,997,884
Number of Sequences: 36976
Number of extensions: 74852
Number of successful extensions: 333
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 327
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 329
length of query: 183
length of database: 9,014,727
effective HSP length: 90
effective length of query: 93
effective length of database: 5,686,887
effective search space: 528880491
effective search space used: 528880491
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0029a.6


