

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0028b.5
(1389 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
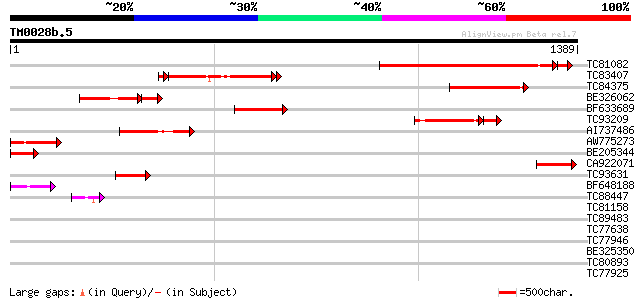
Score E
Sequences producing significant alignments: (bits) Value
TC81082 similar to GP|17385678|dbj|BAB78631. putative co-repress... 670 0.0
TC83407 similar to GP|17385678|dbj|BAB78631. putative co-repress... 320 4e-96
TC84375 similar to GP|17385678|dbj|BAB78631. putative co-repress... 306 3e-83
BE326062 similar to GP|17385678|db putative co-repressor protein... 201 5e-69
BF633689 similar to GP|9945050|gb Similar to Arabidopsis chromos... 240 2e-63
TC93209 similar to PIR|T51447|T51447 transcription regulator-lik... 177 1e-59
AI737486 similar to GP|6094552|gb unknown protein {Arabidopsis t... 166 8e-41
AW775273 similar to PIR|T51447|T5 transcription regulator-like p... 154 2e-37
BE205344 similar to PIR|T00649|T0 hypothetical protein F3I6.12 -... 109 6e-24
CA922071 similar to GP|8247350|emb| pSbaNS5 protein {Sorghum bic... 102 1e-21
TC93631 similar to EGAD|119721|127958 hypothetical protein F54D1... 95 2e-19
BF648188 similar to GP|21903805|gb| hypothetical protein {Strept... 79 9e-15
TC88447 similar to GP|21593015|gb|AAM64964.1 unknown {Arabidopsi... 45 2e-04
TC81158 similar to GP|9280664|gb|AAF86533.1| F21B7.15 {Arabidops... 41 0.003
TC89483 similar to GP|18377662|gb|AAL66981.1 unknown protein {Ar... 40 0.006
TC77638 similar to GP|20161455|dbj|BAB90379. ankyrin-like protei... 40 0.008
TC77946 homologue to GP|10334491|emb|CAC10207. putative splicing... 40 0.008
BE325350 weakly similar to GP|13702821|gb| putative snRNP protei... 39 0.011
TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {... 37 0.070
TC77925 similar to GP|18700163|gb|AAL77693.1 AT4g02720/T10P11_1 ... 37 0.070
>TC81082 similar to GP|17385678|dbj|BAB78631. putative co-repressor protein
{Oryza sativa (japonica cultivar-group)}, partial (20%)
Length = 1461
Score = 670 bits (1728), Expect(2) = 0.0
Identities = 337/435 (77%), Positives = 373/435 (85%)
Frame = +3
Query: 907 QGGDSTRPGTFTNGATTGGTEVHRYQEQSIRHFKSEREEGELSPNGDMEEDNFAVYENSG 966
+GGDS R TNGA GGTEV RYQE+S FKSEREEGELSPNGD EEDNFAVY ++G
Sbjct: 51 EGGDSARLYASTNGAVAGGTEVCRYQEESNPQFKSEREEGELSPNGDFEEDNFAVYGDAG 230
Query: 967 LDAGHKGKGGDMSQKYQSRHGEQVCCEAKGENDVDADDEGEESPHRSSDDSENASENVDV 1026
+A HKG G + ++YQ+R GEQVC EA+GEN DADDEGEESP RSS+ SENAS NVDV
Sbjct: 231 SEAVHKGNDGGLKRQYQNRRGEQVCGEARGENYADADDEGEESPQRSSEGSENASGNVDV 410
Query: 1027 SGSESADGEECSREEHEDREHDNKAESEGEAEGMVDAHDVEGDGMSLTISERFLQTVKPL 1086
SGSESADGEECS+EEH+D EHD+KAESEGEAEGM DAHDVEGDG SL SERFL VKPL
Sbjct: 411 SGSESADGEECSQEEHDDGEHDDKAESEGEAEGMADAHDVEGDGTSLPFSERFLLNVKPL 590
Query: 1087 AKYVPAALHEKDRNSQVFYGSDSFYVLFRLHQTLYERIQSAKFNSSSAEKKWKASHDTGS 1146
AK+VP LH +D+NS+VFYG+DSFYVL RLHQTLYERI +AK NSSS E+KWKAS++T S
Sbjct: 591 AKHVPPVLHVEDKNSRVFYGNDSFYVLIRLHQTLYERIHAAKVNSSSTERKWKASNNTSS 770
Query: 1147 TDQYDRFMNALYSLLDGSSDNTKFEDDCRAIIGTQSYLLFTLDKLIYKLVKQLQAIASEE 1206
TDQYDRFM ALYSLLDGSSDN+KFEDDCRAIIGTQSYLLFTLDKLIYKLVKQLQA+AS+E
Sbjct: 771 TDQYDRFMIALYSLLDGSSDNSKFEDDCRAIIGTQSYLLFTLDKLIYKLVKQLQAVASDE 950
Query: 1207 MDNKLLQLYAYEKSRKLGKFIDIVYHENACVILHEENIYRIEYSPGPKKLSIQLMDCGHD 1266
MDNKLLQLYAYEKSRK GKF+DIVYHENA V+LH+ENIYRIEYSP PK LSIQLMDCG++
Sbjct: 951 MDNKLLQLYAYEKSRKPGKFVDIVYHENARVLLHDENIYRIEYSPKPKTLSIQLMDCGNE 1130
Query: 1267 KPEVTAVSVDPKFSTYLHNDFLSVVPDKKEKSRLFLKRNKHRYAGSSEFSSQAMEGLQVF 1326
K EVTAV VDP FS YLHNDFLS+ DK K +FLKRNK YA S EFSS+AMEGLQ+
Sbjct: 1131 KHEVTAVLVDPNFSAYLHNDFLSIASDK--KLGIFLKRNKRGYACSDEFSSEAMEGLQII 1304
Query: 1327 NGLECKIACSSSKVS 1341
NGLECKIA +SSKVS
Sbjct: 1305 NGLECKIASNSSKVS 1349
Score = 46.6 bits (109), Expect(2) = 0.0
Identities = 22/36 (61%), Positives = 27/36 (74%)
Frame = +1
Query: 1342 YVLDTEDVLYRKKSKRRILHPTSSCNEPAKSSNIRS 1377
YVL+TED L+R +S+R+ LH SSC E K SNIRS
Sbjct: 1351 YVLNTEDFLFRTRSRRKALHLKSSCYEQRKPSNIRS 1458
>TC83407 similar to GP|17385678|dbj|BAB78631. putative co-repressor protein
{Oryza sativa (japonica cultivar-group)}, partial (15%)
Length = 921
Score = 320 bits (820), Expect(3) = 4e-96
Identities = 177/277 (63%), Positives = 210/277 (74%), Gaps = 13/277 (4%)
Frame = +3
Query: 390 VSNKEVSGLKI---KDKHFLKPINELDLSNCDRCTPSYRLLPKNYPIPIASYKSKIGAKV 446
+SNK+VS K+ KDK+ KPINELDLSNC++CTPSYRLLPKNYPIP+AS K+++GAKV
Sbjct: 81 ISNKDVSIPKVSLSKDKYVGKPINELDLSNCEQCTPSYRLLPKNYPIPLASQKTELGAKV 260
Query: 447 LNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDML------LESVNLTTKRVE 500
LNDHWVSVTSGSEDYSFKHMRKNQYEES L+++ ++ V + +
Sbjct: 261 LNDHWVSVTSGSEDYSFKHMRKNQYEESXV*MXKTT-GLNLICC*SL*MQQVRKLKRS*K 437
Query: 501 EL----FEKINNNVIKGDRPVRIDEHLTALNLRCIERLYGDHGLDVMEVLRKNAPLALPV 556
+L F++I V++ + I L ALN + G DVMEVL+KNA LALPV
Sbjct: 438 KLMMI*FQEIFQFVLRSIYQL*I---LGALNDYMVTM-----GFDVMEVLKKNASLALPV 593
Query: 557 ILTRLKQKQDEWARCRADFSKVWAEIYAKNYHKSLDHRSFYFKQQDTKRLSTKALLAEVK 616
ILTRLKQKQ+EWARCR DF+KVWAEIYAKNYHKSLDHRSFYFKQQDTK LSTKALL E+K
Sbjct: 594 ILTRLKQKQEEWARCREDFNKVWAEIYAKNYHKSLDHRSFYFKQQDTKSLSTKALLGEIK 773
Query: 617 EISEKKHKEDDVLLAIAAGSRQPIIPNLEFHYPDLDI 653
EISE+K K DDVLLAIAAG+R+ I+PNLEF YPD +I
Sbjct: 774 EISEQKRKVDDVLLAIAAGNRRXILPNLEFEYPDPEI 884
Score = 45.8 bits (107), Expect(3) = 4e-96
Identities = 20/25 (80%), Positives = 23/25 (92%)
Frame = +2
Query: 364 RDRDRYRDDGMKERDRECRERDRST 388
+DRDR RDDG+KERDRE RERD+ST
Sbjct: 2 KDRDRGRDDGVKERDRELRERDKST 76
Score = 27.3 bits (59), Expect(3) = 4e-96
Identities = 10/11 (90%), Positives = 11/11 (99%)
Frame = +1
Query: 655 EDLYQLVKYSC 665
EDLYQL+KYSC
Sbjct: 889 EDLYQLIKYSC 921
>TC84375 similar to GP|17385678|dbj|BAB78631. putative co-repressor protein
{Oryza sativa (japonica cultivar-group)}, partial (10%)
Length = 587
Score = 306 bits (785), Expect = 3e-83
Identities = 154/197 (78%), Positives = 172/197 (87%), Gaps = 2/197 (1%)
Frame = +3
Query: 1077 ERFLQTVKPLAKYVPAALHEKDRNSQVFYGSDSFYVLFRLHQTLYERIQSAKFNSSSAEK 1136
E FL T KPL KYV H K+ N Q+FYG+D+FYVLFRLHQTLYERI+SAK NSSSAEK
Sbjct: 3 EGFLLTAKPLVKYVSPVFHGKEENVQIFYGNDAFYVLFRLHQTLYERIRSAKINSSSAEK 182
Query: 1137 KWKASHDTGSTDQYDRFMNALYSLLDGSSDNTKFEDDCRAIIGTQSYLLFTLDKLIYKLV 1196
KW+AS+DT STD+Y RFMN+LYSLLDGSSDN+KFEDDCRAIIGTQSY+LFTLDKLIYKLV
Sbjct: 183 KWRASNDTSSTDRYARFMNSLYSLLDGSSDNSKFEDDCRAIIGTQSYVLFTLDKLIYKLV 362
Query: 1197 KQLQAIAS--EEMDNKLLQLYAYEKSRKLGKFIDIVYHENACVILHEENIYRIEYSPGPK 1254
KQLQA+AS +EMDNKLLQLYAYE+SRK G F D+VYHENA V+LH+ENIYRIE S P
Sbjct: 363 KQLQAVASGTDEMDNKLLQLYAYEQSRKSGSFFDVVYHENARVLLHDENIYRIECS--PT 536
Query: 1255 KLSIQLMDCGHDKPEVT 1271
+LSIQLMD GHDKPEVT
Sbjct: 537 RLSIQLMDYGHDKPEVT 587
>BE326062 similar to GP|17385678|db putative co-repressor protein {Oryza
sativa (japonica cultivar-group)}, partial (1%)
Length = 632
Score = 201 bits (510), Expect(2) = 5e-69
Identities = 106/158 (67%), Positives = 122/158 (77%), Gaps = 3/158 (1%)
Frame = +2
Query: 171 DHDRGLLRAEKEQRRRVEKEKDHRED-GKRDWERDGRDHEHDGGQYRERLSHKRKSDRKA 229
DHD G LRAEKE++RR+EKEKD +ED G+R+ ERD RD+EHD G+ RERLSHKRKSD KA
Sbjct: 5 DHDGGSLRAEKERKRRMEKEKDRKEDRGRRERERDYRDYEHDRGRDRERLSHKRKSDHKA 184
Query: 230 EDFGAEPLLDADEFFGTRPMSSTCDDKKSLKTKYPEEFAFCDKVKEKLRNPDGYQEFLKC 289
ED GAEPLLDAD+ FGT+ +E AFCDKVKE+L+NP YQEFLKC
Sbjct: 185 EDSGAEPLLDADQNFGTQ----------------NQELAFCDKVKERLQNPGDYQEFLKC 316
Query: 290 LHIYSKEIITQQELQSLVG--DLLGKYPDLMEEFNEFL 325
+HIY+KEIIT+QELQSLVG LLG YPDLME FNEFL
Sbjct: 317 VHIYNKEIITRQELQSLVGGCGLLGDYPDLMESFNEFL 430
Score = 80.5 bits (197), Expect(2) = 5e-69
Identities = 39/51 (76%), Positives = 41/51 (79%)
Frame = +3
Query: 324 FLVQAEKNDGGFLAGVMNKKSLWIEGPGLKPMKAEDRDRDRDRDRYRDDGM 374
F +QAEKN GGFLAGVMN+KSLWIEG GLKPM AE RDRDRD DR GM
Sbjct: 426 FCLQAEKNGGGFLAGVMNRKSLWIEGRGLKPMNAEQRDRDRDDDRGSVTGM 578
>BF633689 similar to GP|9945050|gb Similar to Arabidopsis chromosome I BAC
genomic sequence (AC002396); unknown protein {Oryza
sativa}, partial (12%)
Length = 398
Score = 240 bits (613), Expect = 2e-63
Identities = 113/130 (86%), Positives = 122/130 (92%)
Frame = +2
Query: 552 LALPVILTRLKQKQDEWARCRADFSKVWAEIYAKNYHKSLDHRSFYFKQQDTKRLSTKAL 611
LALPVILTRLKQKQ+EWARCRADF KVWAEIYAKNYHKSLDHRSFYFKQQDT+ LSTKAL
Sbjct: 8 LALPVILTRLKQKQEEWARCRADFGKVWAEIYAKNYHKSLDHRSFYFKQQDTRSLSTKAL 187
Query: 612 LAEVKEISEKKHKEDDVLLAIAAGSRQPIIPNLEFHYPDLDIHEDLYQLVKYSCGELCTA 671
L E+KEISEKKHKEDDVLLAIAAG+R+PI+PNLEF Y D DIHEDLY+L+KYSC ELCT
Sbjct: 188 LTEIKEISEKKHKEDDVLLAIAAGNRRPILPNLEFEYIDPDIHEDLYRLIKYSCAELCTT 367
Query: 672 EQLDKVMKVW 681
EQLDKVMK+W
Sbjct: 368 EQLDKVMKIW 397
>TC93209 similar to PIR|T51447|T51447 transcription regulator-like protein -
Arabidopsis thaliana, partial (10%)
Length = 721
Score = 177 bits (449), Expect(2) = 1e-59
Identities = 109/175 (62%), Positives = 125/175 (71%), Gaps = 8/175 (4%)
Frame = +3
Query: 993 EAKGENDVDADDEGEESPHRSSDDSENASE-NVDVSGSESADGEECSREEH--EDREHDN 1049
EA G+ND DADDE DSEN SE DVSGSESA G+ECSRE+H ED EHD+
Sbjct: 99 EAGGDNDADADDE----------DSENVSEAGEDVSGSESA-GDECSREDHEEEDMEHDD 245
Query: 1050 ---KAESEGEAEGMVDAHDVEG-DGMSLTISERFLQTVKPLAKYVPAALHEKD-RNSQVF 1104
KAESEGEAEGM DA G DG SL +SERFL TVKPL K+V A +D ++S+VF
Sbjct: 246 VDGKAESEGEAEGMCDADAQTGVDGSSLPLSERFLSTVKPLTKHVSAVSFVEDVKDSRVF 425
Query: 1105 YGSDSFYVLFRLHQTLYERIQSAKFNSSSAEKKWKASHDTGSTDQYDRFMNALYS 1159
YG+D F+VLFRLHQ LYERI SAK NS+SAE KWK + D STD Y RFM+ALY+
Sbjct: 426 YGNDDFFVLFRLHQILYERILSAKENSTSAEIKWK-TKDASSTDLYARFMDALYN 587
Score = 72.8 bits (177), Expect(2) = 1e-59
Identities = 32/44 (72%), Positives = 40/44 (90%)
Frame = +1
Query: 1160 LLDGSSDNTKFEDDCRAIIGTQSYLLFTLDKLIYKLVKQLQAIA 1203
LLDGS +N KFE++CRAI+G QSY+LFTLDKLIYKL++QLQ +A
Sbjct: 589 LLDGSPENAKFENECRAILGNQSYVLFTLDKLIYKLIRQLQTVA 720
>AI737486 similar to GP|6094552|gb unknown protein {Arabidopsis thaliana},
partial (9%)
Length = 473
Score = 166 bits (419), Expect = 8e-41
Identities = 92/183 (50%), Positives = 118/183 (64%)
Frame = +3
Query: 270 CDKVKEKLRNPDGYQEFLKCLHIYSKEIITQQELQSLVGDLLGKYPDLMEEFNEFLVQAE 329
CDKVKEKL + + YQ FLKCL+I+ II + +LQ+LV DLLGK+ DLM EFN+FL + E
Sbjct: 3 CDKVKEKLSSAEDYQTFLKCLNIFGNGIIKKNDLQNLVTDLLGKHSDLMSEFNDFLERCE 182
Query: 330 KNDGGFLAGVMNKKSLWIEGPGLKPMKAEDRDRDRDRDRYRDDGMKERDRECRERDRSTV 389
D GFLAGVM+KK L +G + K ED++ R+ DG KE++R
Sbjct: 183 NID-GFLAGVMSKKPLAGDGHLSRSSKLEDKEHRRE-----TDGGKEKER---------- 314
Query: 390 VSNKEVSGLKIKDKHFLKPINELDLSNCDRCTPSYRLLPKNYPIPIASYKSKIGAKVLND 449
K+K+ K I ELDLS+C RC+PSYRLLP +YPIP AS +S++GA VLND
Sbjct: 315 ----------YKEKYMGKSIQELDLSDCKRCSPSYRLLPADYPIPTASQRSELGAHVLND 464
Query: 450 HWV 452
HWV
Sbjct: 465 HWV 473
>AW775273 similar to PIR|T51447|T5 transcription regulator-like protein -
Arabidopsis thaliana, partial (11%)
Length = 689
Score = 154 bits (390), Expect = 2e-37
Identities = 78/124 (62%), Positives = 96/124 (76%)
Frame = +1
Query: 2 DTAGVIARVKELFEGHRDLILGFNTFLPKGYEITLPLEDEQPPAKKPVEFEEAINFVNKI 61
DT GVIA VKELF+GH +LI GFN +LPKG+EI L ++++ KK VEFE+AI+FV KI
Sbjct: 274 DTHGVIAAVKELFKGHNNLIYGFNAYLPKGHEIRL--DEDEASQKKKVEFEDAISFVGKI 447
Query: 62 KARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHADLLDEFTHFLPDASAAA 121
K RF+ ++HVYKSFLDILNMYR E+K I EVY EVA LF+ H DLL+EFT FLPD S
Sbjct: 448 KNRFQNEEHVYKSFLDILNMYR*EHKTITEVYSEVATLFKDHNDLLEEFTRFLPDNSLEP 627
Query: 122 SSYY 125
S+ +
Sbjct: 628 STQH 639
Score = 35.8 bits (81), Expect = 0.12
Identities = 16/63 (25%), Positives = 33/63 (51%)
Frame = +1
Query: 53 EAINFVNKIKARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHADLLDEFTH 112
+A++++ ++K F Y FL ++ ++ + + V V LF+GH +L+ F
Sbjct: 169 DALSYLKEVKNTFPDQKEKYDMFLQVMKDFKAQKTDTHGVIAAVKELFKGHNNLIYGFNA 348
Query: 113 FLP 115
+LP
Sbjct: 349 YLP 357
>BE205344 similar to PIR|T00649|T0 hypothetical protein F3I6.12 - Arabidopsis
thaliana, partial (8%)
Length = 559
Score = 109 bits (273), Expect = 6e-24
Identities = 52/70 (74%), Positives = 59/70 (84%)
Frame = +2
Query: 1 IDTAGVIARVKELFEGHRDLILGFNTFLPKGYEITLPLEDEQPPAKKPVEFEEAINFVNK 60
+DT GVI RVKELF GHRDLILGFNTFLPKG+EITL EDEQP KK VEF+EA+++VN
Sbjct: 350 VDTDGVIERVKELFRGHRDLILGFNTFLPKGHEITLSSEDEQPQPKKKVEFDEAMSYVNT 529
Query: 61 IKARFEGDDH 70
IK RF+GD H
Sbjct: 530 IKTRFQGDVH 559
Score = 41.2 bits (95), Expect = 0.003
Identities = 21/76 (27%), Positives = 40/76 (52%), Gaps = 2/76 (2%)
Frame = +2
Query: 42 QPPAK--KPVEFEEAINFVNKIKARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVL 99
QPP + + + +A+ ++ +K F+ Y+ FL+++ ++ + + V V L
Sbjct: 209 QPPNRGSQKLTTNDALAYLKAVKDIFQDKKEKYEDFLEVMKDFKAQRVDTDGVIERVKEL 388
Query: 100 FQGHADLLDEFTHFLP 115
F+GH DL+ F FLP
Sbjct: 389 FRGHRDLILGFNTFLP 436
>CA922071 similar to GP|8247350|emb| pSbaNS5 protein {Sorghum bicolor}, partial
(14%)
Length = 808
Score = 102 bits (253), Expect = 1e-21
Identities = 57/97 (58%), Positives = 62/97 (63%)
Frame = -1
Query: 1292 PDKKEKSRLFLKRNKHRYAGSSEFSSQAMEGLQVFNGLECKIACSSSKVSYVLDTEDVLY 1351
P +E K NK + A S E SSQ M+G+QV NGLECKIAC SSKVSYVLDTED L
Sbjct: 778 PTVRESQEFS*KGNKLKCARSDELSSQVMDGIQVINGLECKIACGSSKVSYVLDTEDSLI 599
Query: 1352 RKKSKRRILHPTSSCNEPAKSSNIRSSRALRFCKLFS 1388
R K KR LH +S P SSN SSR RF KLFS
Sbjct: 598 RTKRKRGSLHQNNSYCGPTMSSNTCSSRVQRFYKLFS 488
Score = 38.1 bits (87), Expect = 0.024
Identities = 17/22 (77%), Positives = 18/22 (81%)
Frame = -3
Query: 1283 LHNDFLSVVPDKKEKSRLFLKR 1304
LHNDFLSV PD K KS +FLKR
Sbjct: 806 LHNDFLSVSPDSKRKSGIFLKR 741
>TC93631 similar to EGAD|119721|127958 hypothetical protein F54D1.6
{Caenorhabditis elegans}, partial (1%)
Length = 1081
Score = 95.1 bits (235), Expect = 2e-19
Identities = 44/84 (52%), Positives = 60/84 (71%)
Frame = +1
Query: 260 KTKYPEEFAFCDKVKEKLRNPDGYQEFLKCLHIYSKEIITQQELQSLVGDLLGKYPDLME 319
K + EE F +KVKEK NP+ YQ+FLKCLHIY++E+IT+ ELQ LV DL GKY DLM
Sbjct: 676 KIMFREEHVFLEKVKEKFSNPEIYQDFLKCLHIYARELITRGELQLLVYDLFGKYTDLMV 855
Query: 320 EFNEFLVQAEKNDGGFLAGVMNKK 343
EFN+F+ Q+EK + + + ++
Sbjct: 856 EFNDFMAQSEKKNAKLVKALKGRQ 927
Score = 34.7 bits (78), Expect = 0.26
Identities = 18/36 (50%), Positives = 24/36 (66%), Gaps = 1/36 (2%)
Frame = +3
Query: 284 QEFLKCLHIYSK-EIITQQELQSLVGDLLGKYPDLM 318
+ F K LH Y I ++ELQ +VGDLLGK+ DL+
Sbjct: 501 KNFEKSLHSYRMGTIYKEKELQKMVGDLLGKHSDLV 608
Score = 30.8 bits (68), Expect = 3.8
Identities = 26/90 (28%), Positives = 42/90 (45%), Gaps = 3/90 (3%)
Frame = +1
Query: 256 KKSLKTKYPEEFAFC-DKVKEKLRNPDGYQEFLKCLHIYSK--EIITQQELQSLVGDLLG 312
KK + Y E F ++ ++RN G + L+ +I ++ + ++ +
Sbjct: 412 KKGVSYSYVE*SVFLPEQSHREVRNTRGLPKILRSPYIATEWEQFTRRKNYKKW*AICWE 591
Query: 313 KYPDLMEEFNEFLVQAEKNDGGFLAGVMNK 342
P L E FNEFL Q E N+G F V+NK
Sbjct: 592 NTPILWEGFNEFLTQCESNEGLF-GHVLNK 678
>BF648188 similar to GP|21903805|gb| hypothetical protein {Streptococcus
pyogenes MGAS315}, partial (17%)
Length = 627
Score = 79.3 bits (194), Expect = 9e-15
Identities = 45/112 (40%), Positives = 61/112 (54%)
Frame = +3
Query: 1 IDTAGVIARVKELFEGHRDLILGFNTFLPKGYEITLPLEDEQPPAKKPVEFEEAINFVNK 60
ID GV ARV ELF+GH+ LILG N +PK YEI LP DE+ V ++A F+ +
Sbjct: 309 IDPGGVKARVHELFKGHKHLILGINNIMPKNYEIILPPSDEK------VNRQDATTFLKQ 470
Query: 61 IKARFEGDDHVYKSFLDILNMYRKENKCINEVYHEVAVLFQGHADLLDEFTH 112
+K F+ Y FL +++ Y I +V LF+GH DLL F +
Sbjct: 471 VKVVFQDKMEKYYEFLQVIHDYMNLTIDILDVIEIGMKLFKGHVDLLSGFNY 626
>TC88447 similar to GP|21593015|gb|AAM64964.1 unknown {Arabidopsis
thaliana}, partial (47%)
Length = 1079
Score = 45.1 bits (105), Expect = 2e-04
Identities = 38/100 (38%), Positives = 48/100 (48%), Gaps = 19/100 (19%)
Frame = +2
Query: 151 RERTTVSHCDRDPSVDRP---DPDHDRGLLRAEKEQRRRVEKEKDHREDGKRDWERD--- 204
R+R SH RD DR D D DR LR EK+ R E++ RE +RD +RD
Sbjct: 287 RDRRRDSHRHRDRDYDREYDRDYDRDRYRLREEKDYGR----EREGRERERRDRDRDRGR 454
Query: 205 ----------GRDH-EHDGGQYRERLSHKRKSDRK--AED 231
RD +HDGG YR+R + S R+ AED
Sbjct: 455 RRSYSRSRSRSRDRKDHDGGDYRKRHARSSVSPRRDGAED 574
Score = 34.7 bits (78), Expect = 0.26
Identities = 19/73 (26%), Positives = 37/73 (50%), Gaps = 1/73 (1%)
Frame = +2
Query: 160 DRDPSVDRPDPDHDRGLLRAEKEQRRRVEKEKDHREDGKRDWERD-GRDHEHDGGQYRER 218
D + + P+ D++RG A + R R HR+ RD++R+ RD++ D + RE
Sbjct: 215 DNEQPAEEPEKDYNRGRSPARERDRDRRRDSHRHRD---RDYDREYDRDYDRDRYRLREE 385
Query: 219 LSHKRKSDRKAED 231
+ R+ + + +
Sbjct: 386 KDYGREREGRERE 424
Score = 33.1 bits (74), Expect = 0.77
Identities = 16/28 (57%), Positives = 20/28 (71%)
Frame = +2
Query: 359 DRDRDRDRDRYRDDGMKERDRECRERDR 386
DRD DRDR R R++ R+RE RER+R
Sbjct: 344 DRDYDRDRYRLREEKDYGREREGRERER 427
Score = 32.7 bits (73), Expect = 1.0
Identities = 44/145 (30%), Positives = 66/145 (45%), Gaps = 4/145 (2%)
Frame = +2
Query: 254 DDKKSLKTKYPE-EFAFC--DKVKEKLRNPDGYQEFLKCLHIYSKEIITQQELQSLVGDL 310
+D + L+ K P+ +FA D+V ++L D C I I + L+SL G L
Sbjct: 2 NDYRKLRRKLPDGQFALTHVDEVIDELLTTD-----YSC-DIAMPRIKKRWTLESL-GAL 160
Query: 311 LGKYPDLMEEFNEFLVQAEKNDGGFLAGVMNKKSLWIEGPGLKPMKAEDRDRDRDRDRYR 370
+ L E+F E E+N+ +K G P + DRDR RD R+R
Sbjct: 161 EPRQSALEEDFEE----EEENEDNEQPAEEPEKDY---NRGRSPARERDRDRRRDSHRHR 319
Query: 371 D-DGMKERDRECRERDRSTVVSNKE 394
D D +E DR+ +RDR + K+
Sbjct: 320 DRDYDREYDRD-YDRDRYRLREEKD 391
>TC81158 similar to GP|9280664|gb|AAF86533.1| F21B7.15 {Arabidopsis thaliana},
partial (6%)
Length = 890
Score = 41.2 bits (95), Expect = 0.003
Identities = 23/78 (29%), Positives = 38/78 (48%), Gaps = 2/78 (2%)
Frame = +1
Query: 987 GEQVCCEAKGENDVDADDEGEESPHRSSDDSENASENVDVSGSESADGEECSREEHEDRE 1046
G+ E G D D EGE S +SE+++ + SGS+S + +E E+ E+ +
Sbjct: 370 GDPPSVEVNGNEDKDDGSEGESESESSESESESSTSSSSTSGSDSDEDDEDDAEDEEEGQ 549
Query: 1047 --HDNKAESEGEAEGMVD 1062
D K + A+G +D
Sbjct: 550 ISDDEKMVTWSAADGDLD 603
>TC89483 similar to GP|18377662|gb|AAL66981.1 unknown protein {Arabidopsis
thaliana}, partial (35%)
Length = 1711
Score = 40.0 bits (92), Expect = 0.006
Identities = 29/127 (22%), Positives = 63/127 (48%)
Frame = +1
Query: 150 KRERTTVSHCDRDPSVDRPDPDHDRGLLRAEKEQRRRVEKEKDHREDGKRDWERDGRDHE 209
+R+R +++ ++R + + +R +R E+EQ+RR+E+ + E ++WE R+ E
Sbjct: 142 ERDRERELKREKERVLERYEREAERDRIRKEREQKRRIEEVERQFELQLKEWEYREREKE 321
Query: 210 HDGGQYRERLSHKRKSDRKAEDFGAEPLLDADEFFGTRPMSSTCDDKKSLKTKYPEEFAF 269
+ +E+ + + RK + E D D R + ++K++ + + E+
Sbjct: 322 KERQYEKEKEKDRERKRRKEILYDEE---DDDGDSRKRWRXNAIEEKRNKRLREKED-DL 489
Query: 270 CDKVKEK 276
DK KE+
Sbjct: 490 ADKQKEE 510
>TC77638 similar to GP|20161455|dbj|BAB90379. ankyrin-like protein {Oryza
sativa (japonica cultivar-group)}, partial (62%)
Length = 2345
Score = 39.7 bits (91), Expect = 0.008
Identities = 25/82 (30%), Positives = 47/82 (56%), Gaps = 6/82 (7%)
Frame = +1
Query: 979 SQKYQSRHGEQVCCEAKGENDVDADDEGEE-SPHRSSDDS--ENASENVDVSGSESA--- 1032
+++++ G+ KG+++V ++++ EE S +SS+D+ E+ + + GS +
Sbjct: 526 ARQFEDNPGDLPEDATKGDSNVSSEEKSEENSTEKSSEDTKTEDEGKKTEDEGSNTENNK 705
Query: 1033 DGEECSREEHEDREHDNKAESE 1054
DGEE S +E E E + K ESE
Sbjct: 706 DGEEASTKESESDESEKKDESE 771
Score = 30.0 bits (66), Expect = 6.5
Identities = 33/140 (23%), Positives = 64/140 (45%), Gaps = 1/140 (0%)
Frame = +1
Query: 693 SRAQGAEDTEDVVKAKNTEDV-VNTENNSVKSGIVSVAESGDSKAWQLNGDSGVREDRCL 751
S + +EDT+ + K TED NTENN K G + + +S + +S E+
Sbjct: 619 STEKSSEDTKTEDEGKKTEDEGSNTENN--KDGEEASTKESESDESEKKDES--EENNKS 786
Query: 752 DSDHTVYKTETAGNNTQHGKTNIIGFTPDELSGVNKQNQSSERLVNANVSPASGMEQSNG 811
DSD + K+ + T + + ++ S N ++++ NA ++ + S
Sbjct: 787 DSDESEKKSSDSNETTDSNVEEKVEQSQNKESDENASEKNTDD--NAKDQSSNEVFPSGA 960
Query: 812 RTKIDNTSGHTATPSGNGNT 831
++++ N T T +G+ +T
Sbjct: 961 QSELLN---ETTTQTGSFST 1011
>TC77946 homologue to GP|10334491|emb|CAC10207. putative splicing factor
{Cicer arietinum}, complete
Length = 2061
Score = 39.7 bits (91), Expect = 0.008
Identities = 22/82 (26%), Positives = 47/82 (56%), Gaps = 1/82 (1%)
Frame = +3
Query: 151 RERTTVSHCDRDPSVDRPDPDHDRGLLRAEKEQRRRVEKEKD-HREDGKRDWERDGRDHE 209
RER++ H +RD + + ++ + R +++ R E+EK+ R D +R+ ER+ RD E
Sbjct: 348 RERSSGRHRERDGERGSREKEREKDVERGGRDKERDREREKERERRDKEREKERERRDKE 527
Query: 210 HDGGQYRERLSHKRKSDRKAED 231
+ +ER +++ +R+ ++
Sbjct: 528 RE----KERA*ERKRGERETKE 581
Score = 31.2 bits (69), Expect = 2.9
Identities = 12/28 (42%), Positives = 21/28 (74%)
Frame = +3
Query: 359 DRDRDRDRDRYRDDGMKERDRECRERDR 386
D++RDR+R++ R+ KER++E RD+
Sbjct: 441 DKERDREREKERERRDKEREKERERRDK 524
Score = 30.8 bits (68), Expect = 3.8
Identities = 24/83 (28%), Positives = 35/83 (41%), Gaps = 1/83 (1%)
Frame = +3
Query: 149 VKRERTTVSHCDRDPSVDRPDPDHDRGLLRAEKEQRRRVEKEKDHRE-DGKRDWERDGRD 207
+K ERT H D D D G R K R ++ ++ D RD ERD
Sbjct: 192 IKSERTYRKH--------GIDDDLDGGEDRRSKRSRGGDDENGSKKDRDRDRDRERDREH 347
Query: 208 HEHDGGQYRERLSHKRKSDRKAE 230
E G++RER + +++ E
Sbjct: 348 RERSSGRHRERDGERGSREKERE 416
Score = 30.8 bits (68), Expect = 3.8
Identities = 12/31 (38%), Positives = 22/31 (70%)
Frame = +3
Query: 356 KAEDRDRDRDRDRYRDDGMKERDRECRERDR 386
K DR+R+++R+R + KER+R +ER++
Sbjct: 444 KERDREREKERERRDKEREKERERRDKEREK 536
Score = 29.6 bits (65), Expect = 8.5
Identities = 14/42 (33%), Positives = 27/42 (63%)
Frame = +3
Query: 353 KPMKAEDRDRDRDRDRYRDDGMKERDRECRERDRSTVVSNKE 394
K ++ RD++RDR+R ++ ++++RE +ER+R KE
Sbjct: 417 KDVERGGRDKERDREREKERERRDKERE-KERERRDKEREKE 539
>BE325350 weakly similar to GP|13702821|gb| putative snRNP protein {Oryza
sativa}, partial (6%)
Length = 473
Score = 39.3 bits (90), Expect = 0.011
Identities = 19/34 (55%), Positives = 21/34 (60%)
Frame = +2
Query: 360 RDRDRDRDRYRDDGMKERDRECRERDRSTVVSNK 393
R+RDRDRDR R+ RDRE R RDR NK
Sbjct: 284 RERDRDRDRERERDRDNRDRERRHRDREREEENK 385
Score = 33.9 bits (76), Expect = 0.45
Identities = 23/78 (29%), Positives = 39/78 (49%)
Frame = +2
Query: 154 TTVSHCDRDPSVDRPDPDHDRGLLRAEKEQRRRVEKEKDHREDGKRDWERDGRDHEHDGG 213
+T ++ +R DR D D DRG R +E+D D +R+ +RD RD E
Sbjct: 200 STNNNINRSSDSDRRDRDRDRG-------HDRDSYRERDRDRDRERERDRDNRDRER--- 349
Query: 214 QYRERLSHKRKSDRKAED 231
++R+R +R+ + K +
Sbjct: 350 RHRDR---EREEENKVRE 394
Score = 32.0 bits (71), Expect = 1.7
Identities = 22/64 (34%), Positives = 36/64 (55%), Gaps = 9/64 (14%)
Frame = +2
Query: 357 AEDRDRDRDR----DRYRD-----DGMKERDRECRERDRSTVVSNKEVSGLKIKDKHFLK 407
++ RDRDRDR D YR+ D +ERDR+ R+R+R +E K++++ L+
Sbjct: 233 SDRRDRDRDRGHDRDSYRERDRDRDRERERDRDNRDRERRHRDREREEEN-KVRERARLE 409
Query: 408 PINE 411
+ E
Sbjct: 410 KLAE 421
Score = 31.6 bits (70), Expect = 2.2
Identities = 20/48 (41%), Positives = 28/48 (57%), Gaps = 9/48 (18%)
Frame = +2
Query: 356 KAEDRDRDRDRDRYRDDGMKER---DRE------CRERDRSTVVSNKE 394
+ DRDRDR+R+R RD+ +ER DRE RER R ++ +E
Sbjct: 284 RERDRDRDRERERDRDNRDRERRHRDREREEENKVRERARLEKLAERE 427
>TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {Arabidopsis
thaliana}, partial (39%)
Length = 883
Score = 36.6 bits (83), Expect = 0.070
Identities = 19/61 (31%), Positives = 27/61 (44%)
Frame = +3
Query: 996 GENDVDADDEGEESPHRSSDDSENASENVDVSGSESADGEECSREEHEDREHDNKAESEG 1055
GE + + DEGEE D N + D + DG+E E+ ED E + E +
Sbjct: 378 GEEEDYSGDEGEEEGDPEDDPEANGAGGSDDGEDDDDDGDEEDEEDGEDEEDEEDEEEDD 557
Query: 1056 E 1056
E
Sbjct: 558 E 560
Score = 34.7 bits (78), Expect = 0.26
Identities = 24/74 (32%), Positives = 35/74 (46%), Gaps = 1/74 (1%)
Frame = +3
Query: 997 ENDV-DADDEGEESPHRSSDDSENASENVDVSGSESADGEECSREEHEDREHDNKAESEG 1055
++DV D DD+GEE + S D+ E + D + A G ++ ED + D E E
Sbjct: 348 DDDVQDEDDDGEEEDY-SGDEGEEEGDPEDDPEANGAGGS----DDGEDDDDDGDEEDEE 512
Query: 1056 EAEGMVDAHDVEGD 1069
+ E D D E D
Sbjct: 513 DGEDEEDEEDEEED 554
>TC77925 similar to GP|18700163|gb|AAL77693.1 AT4g02720/T10P11_1
{Arabidopsis thaliana}, partial (47%)
Length = 1538
Score = 36.6 bits (83), Expect = 0.070
Identities = 31/117 (26%), Positives = 48/117 (40%), Gaps = 3/117 (2%)
Frame = +3
Query: 129 RNSVLRDRRSAMPTVRQAHVVKRERTT---VSHCDRDPSVDRPDPDHDRGLLRAEKEQRR 185
R S + R S + ++ V R+R S D D + + D R R + +RR
Sbjct: 603 RKSYKKSRESDSESESESEVEDRKRRKSRKYSESDSDTNSEEEDRKRRR---RRKSSRRR 773
Query: 186 RVEKEKDHREDGKRDWERDGRDHEHDGGQYRERLSHKRKSDRKAEDFGAEPLLDADE 242
R K R + E D D G+ R+R RKS +K+ +EP+ + E
Sbjct: 774 RKSSRKKRRCSDSDESETDDESGYDDSGRKRKRSKRSRKSKKKS----SEPVSEGSE 932
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.133 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 39,825,214
Number of Sequences: 36976
Number of extensions: 546488
Number of successful extensions: 2671
Number of sequences better than 10.0: 92
Number of HSP's better than 10.0 without gapping: 2425
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2578
length of query: 1389
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1281
effective length of database: 5,021,319
effective search space: 6432309639
effective search space used: 6432309639
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0028b.5


