

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
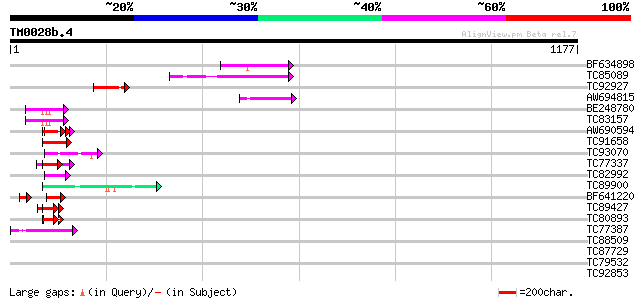
Query= TM0028b.4
(1177 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BF634898 similar to GP|16024936|gb HUA enhancer 2 {Arabidopsis t... 105 8e-23
TC85089 similar to PIR|E96621|E96621 hypothetical protein F23H11... 102 7e-22
TC92927 similar to PIR|E96621|E96621 hypothetical protein F23H11... 79 1e-14
AW694815 similar to GP|22091415|gb putative RNA helicase {Arabid... 60 4e-09
BE248780 similar to GP|5929884|gb|A nucleolin-related protein NR... 52 1e-06
TC83157 similar to GP|21595368|gb|AAM66095.1 unknown {Arabidopsi... 52 1e-06
AW690594 similar to GP|23498163|emb hypothetical protein {Plasmo... 50 7e-06
TC91658 similar to GP|14329812|emb|CAC40753. putative nucleosome... 49 9e-06
TC93070 weakly similar to GP|17065230|gb|AAL32769.1 Unknown prot... 48 2e-05
TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Atel... 46 1e-04
TC82992 homologue to PIR|T05645|T05645 hypothetical protein F20D... 46 1e-04
TC89900 similar to GP|9759355|dbj|BAB10010.1 RNA helicase-like p... 45 2e-04
BF641220 weakly similar to PIR|G86203|G86 probable N-arginine di... 42 3e-04
TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed pr... 44 5e-04
TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {... 44 5e-04
TC77387 similar to GP|15983396|gb|AAL11566.1 At1g51440/F5D21_19 ... 43 6e-04
TC88509 similar to PIR|T01826|T01826 microfibril-associated prot... 42 0.001
TC87729 similar to GP|9759461|dbj|BAB10377.1 gene_id:MAF19.15~un... 42 0.001
TC79532 weakly similar to GP|8164030|gb|AAF73960.1| SGS3 {Arabid... 42 0.001
TC92853 similar to GP|20268748|gb|AAM14077.1 unknown protein {Ar... 42 0.001
>BF634898 similar to GP|16024936|gb HUA enhancer 2 {Arabidopsis thaliana},
partial (21%)
Length = 684
Score = 105 bits (263), Expect = 8e-23
Identities = 61/158 (38%), Positives = 87/158 (54%), Gaps = 5/158 (3%)
Frame = +3
Query: 437 IEPRDMLPAIWFIFSRKGCDAAVQYIEDCKLLDECEKSEVELALKRFRIQYPDAVR---- 492
I R P I F FSR+ C+ + + EK VE + + + R
Sbjct: 84 IMERKFQPVIIFSFSRRECEQHAMSMSKLDFNSQEEKDTVEHVFQNAMLCLNEEDRSLPA 263
Query: 493 -ETAVKGLLQGVAAHHAGCLPLWKAFIEELFQKGLVKVVFATETLAAGINMPARTAVISS 551
E + L +G+A HH+G LP+ K +E LFQ+GLVK +FATET A G+NMPA+T V ++
Sbjct: 264 IELMLPLLQRGIAVHHSGLLPVIKELVELLFQEGLVKALFATETFAMGLNMPAKTVVFTA 443
Query: 552 LSKRIDSGRRPLSSNELLQMAGRAGRRGIDESGHVVLI 589
+ K R + S E +QM+GRAGRRG DE G +++
Sbjct: 444 VKKWDGDSHRYIGSGEYIQMSGRAGRRGKDERGICIIM 557
>TC85089 similar to PIR|E96621|E96621 hypothetical protein F23H11.8
[imported] - Arabidopsis thaliana, partial (24%)
Length = 743
Score = 102 bits (255), Expect = 7e-22
Identities = 81/265 (30%), Positives = 125/265 (46%), Gaps = 8/265 (3%)
Frame = +3
Query: 333 LVTSSKRPVPLTWHF--SMKNSLLPLLDEKGTRMNRKLSLNYLQLQAAGYKPYKDDWSRK 390
+V + RP PL + S L ++DEKG K + Q P D +K
Sbjct: 9 IVYTDYRPTPLQHYIFPSGSEGLYLVVDEKG-----KFREDSFQKALNALIPAADGDRKK 173
Query: 391 RNAR-KRGYRMSYDSDESMFEQRSLSKNNINAIRRSQVPQIIDTLWHIEPRDMLPAIWFI 449
NA+ ++G + ++ES I + I R P I F
Sbjct: 174 ENAKWQKGLVLGKAAEES---------------------DIFKMVKMIIQRQYDPVILFN 290
Query: 450 FSRKGCDAAVQYIEDCKLLDECEKSEVE-LALKRFRIQYPDAVRETAVKGLL----QGVA 504
FS++ C+ + L + EK +E + + D + V +L +G+
Sbjct: 291 FSKRECEFLAMQMAKMDLNGDIEKDNIEKIFWCAMDMLSDDDKKLPQVSNMLPLLKRGIG 470
Query: 505 AHHAGCLPLWKAFIEELFQKGLVKVVFATETLAAGINMPARTAVISSLSKRIDSGRRPLS 564
HH+G LP+ K IE LFQ+GL+K +FATET + G+NMPA+T V +++ K R ++
Sbjct: 471 VHHSGLLPILKEVIEILFQEGLIKCLFATETFSIGLNMPAKTVVFTNVRKFDGDKFRWIT 650
Query: 565 SNELLQMAGRAGRRGIDESGHVVLI 589
S E +QM+GRAGRRGID+ G +L+
Sbjct: 651 SGEYIQMSGRAGRRGIDDRGVCILM 725
>TC92927 similar to PIR|E96621|E96621 hypothetical protein F23H11.8
[imported] - Arabidopsis thaliana, partial (11%)
Length = 655
Score = 78.6 bits (192), Expect = 1e-14
Identities = 39/75 (52%), Positives = 56/75 (74%)
Frame = +3
Query: 175 SVVVSAPTSSGKTLIAEAAAVATVARGRRIFYTTPLKALSNQKFREFRETFGDSYVGLLT 234
SV+VSA TS+GKT++A A ++ +R+ YT+P+KALSNQK+REF+E F D VGL+T
Sbjct: 435 SVMVSAHTSAGKTVVALYAIAMSLRNKQRVIYTSPIKALSNQKYREFKEEFSD--VGLMT 608
Query: 235 GDSAVNKDAQVLIMT 249
GD ++ +A L+MT
Sbjct: 609 GDVTIDPNASCLVMT 653
>AW694815 similar to GP|22091415|gb putative RNA helicase {Arabidopsis
thaliana}, partial (16%)
Length = 404
Score = 60.5 bits (145), Expect = 4e-09
Identities = 42/122 (34%), Positives = 61/122 (49%), Gaps = 4/122 (3%)
Frame = +3
Query: 477 ELALKRFRIQYPDA-VRETAVKGLLQGVAAHHAGCLPLWKAFIEELFQKGLVKVVFATET 535
E AL+ F Q D +R T L G+ HHAG ++ +EELF ++V+ T T
Sbjct: 42 EEALEMFLSQVSDQNLRHT----LQFGIGLHHAGLNDKDRSLVEELFANNKIQVLVCTST 209
Query: 536 LAAGINMPARTAVISSLSKRIDSGRR--PLSSNELLQMAGRAGRRGIDESGH-VVLIQTP 592
LA G+N+PA +I +R ++LQM GRAGR D+ G V+L+ P
Sbjct: 210 LAWGVNLPAHLVIIKGTEYYDGKTKRYVDFPITDILQMMGRAGRPQFDQHGKAVILVHEP 389
Query: 593 NE 594
+
Sbjct: 390 KK 395
>BE248780 similar to GP|5929884|gb|A nucleolin-related protein NRP {Rattus
norvegicus}, partial (4%)
Length = 561
Score = 52.0 bits (123), Expect = 1e-06
Identities = 43/121 (35%), Positives = 60/121 (49%), Gaps = 32/121 (26%)
Frame = +2
Query: 33 PTTTL---PLSLRSLSST--LKFRLSFSFNPPTSSSLPT-----------FSDAGEDD-- 74
PT +L PLSLR LSS L +S P ++S PT FS + +DD
Sbjct: 128 PTLSLFKDPLSLRLLSSAALLNHNISSPLLPIFTNSKPTSAFGQWHHGRYFSSSKQDDHI 307
Query: 75 -------VEDEDDD-----DEEEEEYEEEE--YEDDDDDDVAADEYDDEVPDEGEAFDAS 120
+EDEDDD D+++++Y+EEE YED+DDD VA +E + +A
Sbjct: 308 KEGGTHEIEDEDDDSDGDDDDDDDDYDEEEGDYEDEDDDTVAVSSRKKVYTEEEKEAEAE 487
Query: 121 A 121
A
Sbjct: 488 A 490
>TC83157 similar to GP|21595368|gb|AAM66095.1 unknown {Arabidopsis
thaliana}, partial (57%)
Length = 1108
Score = 52.0 bits (123), Expect = 1e-06
Identities = 43/121 (35%), Positives = 60/121 (49%), Gaps = 32/121 (26%)
Frame = +3
Query: 33 PTTTL---PLSLRSLSST--LKFRLSFSFNPPTSSSLPT-----------FSDAGEDD-- 74
PT +L PLSLR LSS L +S P ++S PT FS + +DD
Sbjct: 135 PTLSLFKDPLSLRLLSSAALLNHNISSPLLPIFTNSKPTSAFGQWHHGRYFSSSKQDDHI 314
Query: 75 -------VEDEDDD-----DEEEEEYEEEE--YEDDDDDDVAADEYDDEVPDEGEAFDAS 120
+EDEDDD D+++++Y+EEE YED+DDD VA +E + +A
Sbjct: 315 KEGGTHEIEDEDDDSDGDDDDDDDDYDEEEGDYEDEDDDTVAVSSRKKVYTEEEKEAEAE 494
Query: 121 A 121
A
Sbjct: 495 A 497
>AW690594 similar to GP|23498163|emb hypothetical protein {Plasmodium
falciparum 3D7}, partial (10%)
Length = 633
Score = 49.7 bits (117), Expect = 7e-06
Identities = 28/63 (44%), Positives = 37/63 (58%)
Frame = +3
Query: 72 EDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGEAFDASARREEFKWQRV 131
E D E+ED+ DEEEEE EEEE ED+ DD +E ++E +E E D EE + R+
Sbjct: 96 EHDEEEEDEHDEEEEEEEEEEEEDEHDD----EEEEEEEEEEEEEEDDDEEGEEDEIDRI 263
Query: 132 EKL 134
L
Sbjct: 264 STL 272
Score = 48.1 bits (113), Expect = 2e-05
Identities = 21/47 (44%), Positives = 33/47 (69%)
Frame = +3
Query: 69 DAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGE 115
D +++ EDE D++EEEEE EEEE E DD+++ +E ++E D+ E
Sbjct: 93 DEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEEEEEEEEEEEEEDDDE 233
Score = 47.0 bits (110), Expect = 4e-05
Identities = 20/47 (42%), Positives = 36/47 (76%)
Frame = +3
Query: 69 DAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGE 115
D GE + E+++ D+EEE+E++EEE E++++++ DE+DDE +E E
Sbjct: 66 DEGEVEEEEDEHDEEEEDEHDEEEEEEEEEEE--EDEHDDEEEEEEE 200
Score = 46.6 bits (109), Expect = 6e-05
Identities = 20/54 (37%), Positives = 35/54 (64%)
Frame = +3
Query: 72 EDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGEAFDASARREE 125
ED+ ++E++D+ +EEE EEEE E++D+ D +E ++E +E E D +E
Sbjct: 90 EDEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEEEEEEEEEEEEEDDDEEGEEDE 251
Score = 40.8 bits (94), Expect = 0.003
Identities = 19/54 (35%), Positives = 32/54 (59%)
Frame = +3
Query: 72 EDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGEAFDASARREE 125
E+ E+ED+ + EEEE E +E E+D+ D+ +E ++E DE + + EE
Sbjct: 45 EEHNEEEDEGEVEEEEDEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEEEEEEEE 206
Score = 39.7 bits (91), Expect = 0.007
Identities = 19/55 (34%), Positives = 35/55 (63%), Gaps = 2/55 (3%)
Frame = +3
Query: 73 DDVEDEDDDDEEEEEYEEEEYE--DDDDDDVAADEYDDEVPDEGEAFDASARREE 125
++ EDE + +EEE+E++EEE + D+++++ +E +DE DE E + EE
Sbjct: 54 NEEEDEGEVEEEEDEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEEEEEEEEEEEE 218
Score = 35.8 bits (81), Expect = 0.100
Identities = 18/57 (31%), Positives = 35/57 (60%)
Frame = +3
Query: 77 DEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGEAFDASARREEFKWQRVEK 133
+E +++E+E E EEEE E D++++ DE+D+E +E E + +E + + E+
Sbjct: 45 EEHNEEEDEGEVEEEEDEHDEEEE---DEHDEEEEEEEEEEEEDEHDDEEEEEEEEE 206
>TC91658 similar to GP|14329812|emb|CAC40753. putative nucleosome assembly
protein 1 {Atropa belladonna}, partial (46%)
Length = 583
Score = 49.3 bits (116), Expect = 9e-06
Identities = 25/59 (42%), Positives = 38/59 (64%)
Frame = +2
Query: 69 DAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGEAFDASARREEFK 127
+AGE D +D + D E++E+ +EE+ +DDDDDD ++ DDE DEG+ S R + K
Sbjct: 50 EAGESDFDDIEVD-EDDEDGDEEDDDDDDDDDDDEEDEDDEEEDEGKGKSKSKRGSKAK 223
Score = 29.6 bits (65), Expect = 7.2
Identities = 13/49 (26%), Positives = 26/49 (52%)
Frame = +2
Query: 85 EEEYEEEEYEDDDDDDVAADEYDDEVPDEGEAFDASARREEFKWQRVEK 133
E ++++ E ++DD+D D+ DD+ D+ E + +E K + K
Sbjct: 59 ESDFDDIEVDEDDEDGDEEDDDDDDDDDDDEEDEDDEEEDEGKGKSKSK 205
>TC93070 weakly similar to GP|17065230|gb|AAL32769.1 Unknown protein
{Arabidopsis thaliana}, partial (19%)
Length = 679
Score = 48.1 bits (113), Expect = 2e-05
Identities = 38/126 (30%), Positives = 66/126 (52%), Gaps = 6/126 (4%)
Frame = +1
Query: 72 EDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGEAFDASARREEFKWQRV 131
+D++E+ D +D+E+E+ +E+E +DDDDDD D+ DD+ E D +A+ + K + V
Sbjct: 226 DDEIEEMDFEDDEDEDEDEDEDDDDDDDD---DDEDDDDDGFAEPTDLNAKDKRLKSKTV 396
Query: 132 -EKLCNEVREFGAEIIDVDELASVYDFRIDKFQRLAV-----QAFLRGSSVVVSAPTSSG 185
++ + +E G V L + R+ K QR+A A R +S+ P S
Sbjct: 397 PDRQQEKEKEKG-----VRSLNNGQSKRLPKSQRIASLQENNSAKFRRNSMEKKYPELSE 561
Query: 186 KTLIAE 191
+ L+ E
Sbjct: 562 EILLDE 579
>TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Ateline
herpesvirus 3}, partial (8%)
Length = 986
Score = 45.8 bits (107), Expect = 1e-04
Identities = 22/41 (53%), Positives = 28/41 (67%)
Frame = +3
Query: 69 DAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDE 109
D EDD EDEDDDDEEE E+EE D++D+ +E +DE
Sbjct: 621 DQDEDDDEDEDDDDEEEGGEEDEEEGVDEEDNEEEEEDEDE 743
Score = 43.9 bits (102), Expect = 4e-04
Identities = 24/80 (30%), Positives = 43/80 (53%), Gaps = 1/80 (1%)
Frame = +3
Query: 55 SFNPPTSSSLPTFSDAGEDDV-EDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDE 113
S N S P G D+ E+EDD+D ++++ +++E EDDDD++ +E ++E DE
Sbjct: 528 SNNKSNSKKAPEGGAGGADENGEEEDDEDGDDQDEDDDEDEDDDDEEEGGEEDEEEGVDE 707
Query: 114 GEAFDASARREEFKWQRVEK 133
+ + +E Q +K
Sbjct: 708 EDNEEEEEDEDEEALQPPKK 767
Score = 33.1 bits (74), Expect = 0.65
Identities = 20/88 (22%), Positives = 38/88 (42%)
Frame = +3
Query: 62 SSLPTFSDAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGEAFDASA 121
+ +PT + +D+ D E E+ ++E+ EDDD D + D+ ++G + ++
Sbjct: 351 TDMPTTKGDSKTQSQDKQHDAEGEDGNDDEDEEDDDGDGAFGEGEDELSSEDGGGYGNNS 530
Query: 122 RREEFKWQRVEKLCNEVREFGAEIIDVD 149
+ + E E G E D D
Sbjct: 531 NNKSNSKKAPEGGAGGADENGEEEDDED 614
>TC82992 homologue to PIR|T05645|T05645 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (7%)
Length = 354
Score = 45.8 bits (107), Expect = 1e-04
Identities = 25/59 (42%), Positives = 34/59 (57%), Gaps = 4/59 (6%)
Frame = +1
Query: 72 EDDVEDEDDDDEEEEEYEEEEYEDDDDDD----VAADEYDDEVPDEGEAFDASARREEF 126
E++ E D++EEEEYEEEE E++DDDD V E D P EG F++ + F
Sbjct: 64 EEEARIEMMDEDEEEEYEEEEEEEEDDDDGSGAVYFPEAGDLEPSEGMEFESEEAAKAF 240
>TC89900 similar to GP|9759355|dbj|BAB10010.1 RNA helicase-like protein
{Arabidopsis thaliana}, partial (49%)
Length = 1515
Score = 44.7 bits (104), Expect = 2e-04
Identities = 63/274 (22%), Positives = 111/274 (39%), Gaps = 27/274 (9%)
Frame = +1
Query: 68 SDAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGEAFDA--SARREE 125
SD G DD+ E + + + + ++DD++ +AA+ + +V + G+
Sbjct: 112 SDDG-DDLRMEKPNKKNSKRKVQLSNDNDDNEVIAAEVGNLKVVNGGDVSSGIVGVTASY 288
Query: 126 FKWQRVEKLCNEVREFGAEIIDVDELASVYDFRIDKF---QRLAVQAFLRGSSVVVSAPT 182
R +K C+ + L V D +K Q + L+G V+ A T
Sbjct: 289 LSDSRFDK-CS---------VSPLSLKGVKDAGYEKMTIVQEATLPVILKGKDVLAKAKT 438
Query: 183 SSGKTLIAEAAAVATVA------RGRR-----IFYTTPLKALSNQ------KFREFRETF 225
+GKT+ ++ V RG+ + P + L+ Q K ++ T
Sbjct: 439 GTGKTVAFLLPSIEVVVKSPPNNRGQNQTPILVLVVCPTRELACQAAAEATKLLKYHPTI 618
Query: 226 GDSYV----GLLTGDSAVNKD-AQVLIMTTEILRNMLYQSVGNVSSSRSGLVNVDVIVLD 280
G V L + + K+ Q+LI T L++ + N + S L+ V +VLD
Sbjct: 619 GAQVVIGGTSLSSEQKRIQKNPCQILIATPGRLKD----HIANTAGFASKLMGVKALVLD 786
Query: 281 EVHYLSDISRGTVWEEIVIYCPKEVQLICLSATV 314
E L D+ E+I+ PK+ Q + SAT+
Sbjct: 787 EADQLLDMGFRKDIEKIIASVPKQRQTLMFSATI 888
>BF641220 weakly similar to PIR|G86203|G86 probable N-arginine dibasic
convertase [imported] - Arabidopsis thaliana, partial
(5%)
Length = 634
Score = 42.0 bits (97), Expect(2) = 3e-04
Identities = 17/40 (42%), Positives = 28/40 (69%)
Frame = +2
Query: 76 EDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGE 115
+D D+++EEE +E+E +D++DDD D+ D+E DE E
Sbjct: 344 KDGSIDEDDEEEDDEDEEDDEEDDDEGEDDEDEEXEDEDE 463
Score = 41.6 bits (96), Expect = 0.002
Identities = 16/38 (42%), Positives = 28/38 (73%)
Frame = +2
Query: 71 GEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDD 108
G D +DE++DDE+EE+ EE++ E +DD+D ++ D+
Sbjct: 350 GSIDEDDEEEDDEDEEDDEEDDDEGEDDEDEEXEDEDE 463
Score = 40.4 bits (93), Expect = 0.004
Identities = 14/29 (48%), Positives = 26/29 (89%)
Frame = +2
Query: 72 EDDVEDEDDDDEEEEEYEEEEYEDDDDDD 100
++D E+EDD+DEE++E +++E EDD+D++
Sbjct: 359 DEDDEEEDDEDEEDDEEDDDEGEDDEDEE 445
Score = 39.3 bits (90), Expect = 0.009
Identities = 15/32 (46%), Positives = 26/32 (80%)
Frame = +2
Query: 69 DAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDD 100
D E+D EDE+DD+E+++E E++E E+ +D+D
Sbjct: 365 DDEEEDDEDEEDDEEDDDEGEDDEDEEXEDED 460
Score = 38.5 bits (88), Expect = 0.015
Identities = 15/34 (44%), Positives = 25/34 (73%)
Frame = +2
Query: 72 EDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADE 105
++D ED+++DD+E E+ E+EE ED+D+ V E
Sbjct: 383 DEDEEDDEEDDDEGEDDEDEEXEDEDEXXVXGRE 484
Score = 38.1 bits (87), Expect = 0.020
Identities = 15/41 (36%), Positives = 27/41 (65%)
Frame = +2
Query: 65 PTFSDAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADE 105
P EDD E++D+D+E++EE ++E +D+D++ DE
Sbjct: 341 PKDGSIDEDDEEEDDEDEEDDEEDDDEGEDDEDEEXEDEDE 463
Score = 38.1 bits (87), Expect = 0.020
Identities = 14/42 (33%), Positives = 29/42 (68%)
Frame = +2
Query: 69 DAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEV 110
D D+ ++E+DD++EE++ E+++ +DD+D+ DE + V
Sbjct: 347 DGSIDEDDEEEDDEDEEDDEEDDDEGEDDEDEEXEDEDEXXV 472
Score = 32.7 bits (73), Expect = 0.85
Identities = 16/43 (37%), Positives = 25/43 (57%)
Frame = +2
Query: 76 EDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGEAFD 118
E D +E+ EEE+ ED++DD+ DE +D+ +E E D
Sbjct: 332 EGAPKDGSIDEDDEEEDDEDEEDDEEDDDEGEDDEDEEXEDED 460
Score = 21.2 bits (43), Expect(2) = 3e-04
Identities = 12/25 (48%), Positives = 15/25 (60%)
Frame = +3
Query: 20 PPRRYPSFSLLPIPTTTLPLSLRSL 44
PP R P LL + TTL SLR++
Sbjct: 183 PPPRPPLPRLLCLHLTTLLSSLRTI 257
>TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed protein
{Arabidopsis thaliana}, partial (50%)
Length = 753
Score = 43.5 bits (101), Expect = 5e-04
Identities = 15/42 (35%), Positives = 29/42 (68%)
Frame = +1
Query: 59 PTSSSLPTFSDAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDD 100
P +P + +DD ED+DDDD++ ++ E+E+ ++++DDD
Sbjct: 409 PEDDPVPNGAGGSDDDDEDDDDDDDDNDDGEDEDEDEEEDDD 534
Score = 43.1 bits (100), Expect = 6e-04
Identities = 19/45 (42%), Positives = 31/45 (68%)
Frame = +1
Query: 68 SDAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPD 112
S+ +DD ED+DDD +E++ +E++ D+DD+ AD DD VP+
Sbjct: 301 SETEDDDDEDDDDDVNDEDDDNDEDFSGDEDDE-DADPEDDPVPN 432
Score = 42.0 bits (97), Expect = 0.001
Identities = 24/66 (36%), Positives = 36/66 (54%), Gaps = 12/66 (18%)
Frame = +1
Query: 72 EDDVEDEDDDDEEE----EEYEEEEYEDD--------DDDDVAADEYDDEVPDEGEAFDA 119
+DDV DEDDD++E+ E+ E+ + EDD DDD D+ DD+ D+GE D
Sbjct: 334 DDDVNDEDDDNDEDFSGDEDDEDADPEDDPVPNGAGGSDDDDEDDDDDDDDNDDGEDEDE 513
Query: 120 SARREE 125
++
Sbjct: 514 DEEEDD 531
Score = 40.0 bits (92), Expect = 0.005
Identities = 17/47 (36%), Positives = 31/47 (65%)
Frame = +1
Query: 69 DAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGE 115
+ +D E EDDDDE++++ +E +DD+D+D + DE D++ E +
Sbjct: 283 EENKDGSETEDDDDEDDDDDVNDE-DDDNDEDFSGDEDDEDADPEDD 420
Score = 39.7 bits (91), Expect = 0.007
Identities = 18/40 (45%), Positives = 28/40 (70%), Gaps = 4/40 (10%)
Frame = +1
Query: 65 PTFSDAGEDDVEDEDDDDEEEE----EYEEEEYEDDDDDD 100
P + AG D +DEDDDD++++ E E+E+ E+DDD+D
Sbjct: 421 PVPNGAGGSDDDDEDDDDDDDDNDDGEDEDEDEEEDDDED 540
Score = 38.9 bits (89), Expect = 0.012
Identities = 23/58 (39%), Positives = 31/58 (52%), Gaps = 8/58 (13%)
Frame = +1
Query: 69 DAGEDDVEDEDDDDEEEEE--------YEEEEYEDDDDDDVAADEYDDEVPDEGEAFD 118
D ED DEDD+D + E+ +++ EDDDDDD D+ +DE DE E D
Sbjct: 361 DNDEDFSGDEDDEDADPEDDPVPNGAGGSDDDDEDDDDDDDDNDDGEDEDEDEEEDDD 534
Score = 35.4 bits (80), Expect = 0.13
Identities = 21/68 (30%), Positives = 35/68 (50%), Gaps = 13/68 (19%)
Frame = +1
Query: 70 AGEDDVEDED-------------DDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGEA 116
+G++D ED D DDD+E+++ +DDDD+D DE +DE D+ E
Sbjct: 379 SGDEDDEDADPEDDPVPNGAGGSDDDDEDDD------DDDDDNDDGEDEDEDEEEDDDED 540
Query: 117 FDASARRE 124
S +++
Sbjct: 541 QPPSKKKK 564
Score = 29.3 bits (64), Expect = 9.3
Identities = 10/19 (52%), Positives = 17/19 (88%)
Frame = +1
Query: 68 SDAGEDDVEDEDDDDEEEE 86
+D GED+ EDE++DD+E++
Sbjct: 487 NDDGEDEDEDEEEDDDEDQ 543
>TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {Arabidopsis
thaliana}, partial (39%)
Length = 883
Score = 43.5 bits (101), Expect = 5e-04
Identities = 20/41 (48%), Positives = 28/41 (67%)
Frame = +3
Query: 71 GEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVP 111
G DD ED+DDD +EE+E + E+ ED++D +E DDE P
Sbjct: 459 GSDDGEDDDDDGDEEDEEDGEDEEDEED-----EEEDDETP 566
Score = 42.7 bits (99), Expect = 8e-04
Identities = 17/33 (51%), Positives = 27/33 (81%)
Frame = +3
Query: 68 SDAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDD 100
SD GEDD +D D++DEE+ E EE+E ++++DD+
Sbjct: 462 SDDGEDDDDDGDEEDEEDGEDEEDEEDEEEDDE 560
Score = 41.6 bits (96), Expect = 0.002
Identities = 26/66 (39%), Positives = 35/66 (52%), Gaps = 9/66 (13%)
Frame = +3
Query: 69 DAGEDDVEDEDDDDEEEE-EYEEEEYEDDDDDDVAAD--------EYDDEVPDEGEAFDA 119
D +DDV+DEDDD EEE+ +E E E D +DD A+ E DD+ DE + D
Sbjct: 339 DDDDDDVQDEDDDGEEEDYSGDEGEEEGDPEDDPEANGAGGSDDGEDDDDDGDEEDEEDG 518
Query: 120 SARREE 125
+E
Sbjct: 519 EDEEDE 536
Score = 40.4 bits (93), Expect = 0.004
Identities = 22/61 (36%), Positives = 34/61 (55%), Gaps = 7/61 (11%)
Frame = +3
Query: 72 EDDVEDEDDDD--EEEEEYEEEEYEDDDDDDVAADEYDDEV-----PDEGEAFDASARRE 124
EDD +D+DDDD +E+++ EEE+Y D+ ++ E D E D+GE D E
Sbjct: 324 EDDEDDDDDDDVQDEDDDGEEEDYSGDEGEEEGDPEDDPEANGAGGSDDGEDDDDDGDEE 503
Query: 125 E 125
+
Sbjct: 504 D 506
>TC77387 similar to GP|15983396|gb|AAL11566.1 At1g51440/F5D21_19
{Arabidopsis thaliana}, partial (75%)
Length = 1793
Score = 43.1 bits (100), Expect = 6e-04
Identities = 43/150 (28%), Positives = 71/150 (46%), Gaps = 9/150 (6%)
Frame = +3
Query: 1 MAFVATTNTPPLILSHSLRPPRRYPSFSLLPIPTTTLPLSLRS----LSSTLKFRLSFSF 56
MA V P + HS+ P YPS S + P++ +S LS++ K S
Sbjct: 117 MASVVPVLLPNTLNQHSILP---YPSHSH---HLFSKPINHKSNILFLSNSKKPNNSPIL 278
Query: 57 NPPTSSSLPTFSDAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAAD---EYDDEVPDE 113
SSS T + E++ E+E++D++EEEE EEEE E++ ++ E E E
Sbjct: 279 FLKCSSSSSTMNQLHEEEEEEEEEDEDEEEEEEEEEEEEESPLPPLSEVWREIQGENDWE 458
Query: 114 G--EAFDASARREEFKWQRVEKLCNEVREF 141
G + D R+E ++ + + C + +F
Sbjct: 459 GLLDPMDPILRKEIIRYGELAQACYDSFDF 548
>TC88509 similar to PIR|T01826|T01826 microfibril-associated protein homolog
T15F16.8 - Arabidopsis thaliana, partial (83%)
Length = 1306
Score = 42.4 bits (98), Expect = 0.001
Identities = 27/85 (31%), Positives = 45/85 (52%), Gaps = 10/85 (11%)
Frame = +1
Query: 76 EDEDDDDEEEEEYEEEEYEDDDDDD---------VAADEYDDEVPDEGEAFDASARR-EE 125
E+E++++EEEEE EE +YE D D++ V + + + E E +A EE
Sbjct: 502 EEEEEEEEEEEEEEESDYETDSDEEYTGVAMVKPVFVPKSERDTIAERERLEAEEEALEE 681
Query: 126 FKWQRVEKLCNEVREFGAEIIDVDE 150
+ +R+E+ NE R+ E I D+
Sbjct: 682 ARKRRMEERRNETRQIVVEEIRKDQ 756
>TC87729 similar to GP|9759461|dbj|BAB10377.1 gene_id:MAF19.15~unknown protein
{Arabidopsis thaliana}, partial (37%)
Length = 2210
Score = 42.4 bits (98), Expect = 0.001
Identities = 29/72 (40%), Positives = 37/72 (51%), Gaps = 12/72 (16%)
Frame = +3
Query: 55 SFNPPTSSSLPT--FSDAGEDDVEDEDDDDEEEE--------EYEEEEYEDDDDDDVAAD 104
SF PP SS + D D+ E E +DDEEEE + E EYED+++D+
Sbjct: 1551 SFLPPAKSSRHQVGYPDDERDESEYETEDDEEEERPRSRMRDDDSEAEYEDEEEDEQIEQ 1730
Query: 105 EYD--DEVPDEG 114
E D DE DEG
Sbjct: 1731 ENDASDEDEDEG 1766
Score = 35.0 bits (79), Expect = 0.17
Identities = 18/62 (29%), Positives = 34/62 (54%), Gaps = 7/62 (11%)
Frame = +3
Query: 72 EDDVEDE-------DDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGEAFDASARRE 124
EDD E+E DDD E E E EEE+ + + ++D + ++ D+ + + + A+++
Sbjct: 1632 EDDEEEERPRSRMRDDDSEAEYEDEEEDEQIEQENDASDEDEDEGLKQKSKNLKRKAKKK 1811
Query: 125 EF 126
F
Sbjct: 1812 GF 1817
>TC79532 weakly similar to GP|8164030|gb|AAF73960.1| SGS3 {Arabidopsis
thaliana}, partial (47%)
Length = 1558
Score = 42.4 bits (98), Expect = 0.001
Identities = 20/57 (35%), Positives = 34/57 (59%)
Frame = +1
Query: 72 EDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGEAFDASARREEFKW 128
E + E+DDD+EEEE ++ + +D DDD+ +DEYD + + + R++ KW
Sbjct: 640 EQKNDGEEDDDDEEEEEDDCDDLEDTDDDLMSDEYDSDASQK-----SHETRKKSKW 795
>TC92853 similar to GP|20268748|gb|AAM14077.1 unknown protein {Arabidopsis
thaliana}, partial (21%)
Length = 761
Score = 42.0 bits (97), Expect = 0.001
Identities = 24/83 (28%), Positives = 44/83 (52%), Gaps = 2/83 (2%)
Frame = +3
Query: 39 LSLRSLSSTLKFRLSFSFNPPT--SSSLPTFSDAGEDDVEDEDDDDEEEEEYEEEEYEDD 96
+ + ++ S+ + R + SF P+ +P + + D D+DDE +E EEEE ED+
Sbjct: 237 IDMSNIVSSTRRRATISFVAPSPPKPKIPVETSGKDTKGSDNDNDDEGKENEEEEEDEDE 416
Query: 97 DDDDVAADEYDDEVPDEGEAFDA 119
+++D + D+ E E F+A
Sbjct: 417 EEEDTSDDD-----GSESEEFNA 470
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.134 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,470,477
Number of Sequences: 36976
Number of extensions: 467291
Number of successful extensions: 7339
Number of sequences better than 10.0: 263
Number of HSP's better than 10.0 without gapping: 3747
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5306
length of query: 1177
length of database: 9,014,727
effective HSP length: 107
effective length of query: 1070
effective length of database: 5,058,295
effective search space: 5412375650
effective search space used: 5412375650
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0028b.4


