

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0028b.3
(288 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
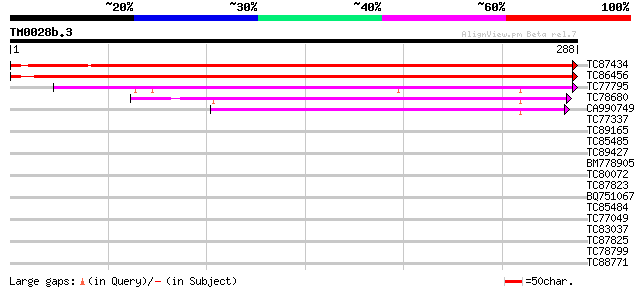
Score E
Sequences producing significant alignments: (bits) Value
TC87434 similar to SP|P52902|ODPA_PEA Pyruvate dehydrogenase E1 ... 482 e-137
TC86456 similar to SP|P52902|ODPA_PEA Pyruvate dehydrogenase E1 ... 464 e-131
TC77795 similar to GP|15450707|gb|AAK96625.1 At1g01090/T25K16_8 ... 180 5e-46
TC78680 similar to GP|9454571|gb|AAF87894.1| branched-chain alph... 96 2e-20
CA990749 similar to GP|9955517|emb branched-chain alpha keto-aci... 94 6e-20
TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Atel... 31 0.63
TC89165 similar to PIR|S19652|S19652 cellodextrinase C - Pseudom... 30 0.82
TC85485 similar to GP|22136900|gb|AAM91794.1 putative transketol... 30 1.4
TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed pr... 29 2.4
BM778905 similar to SP|Q05046|CH62_ Chaperonin CPN60-2 mitochon... 28 3.1
TC80072 similar to GP|14423520|gb|AAK62442.1 Unknown protein {Ar... 28 4.1
TC87823 similar to GP|9295275|gb|AAF86907.1| phosphoenolpyruvate... 28 4.1
BQ751067 similar to GP|18461169|dbj hypothetical protein~similar... 28 5.3
TC85484 similar to PIR|S58083|S58083 transketolase (EC 2.2.1.1) ... 27 7.0
TC77049 homologue to GP|3941289|gb|AAC82326.1| similarity to SCA... 27 7.0
TC83037 weakly similar to PIR|T38758|T38758 60s ribosomal protei... 27 7.0
TC87825 similar to GP|21553995|gb|AAM63076.1 PP2A regulatory sub... 27 7.0
TC78799 weakly similar to SP|P96132|THIO_THIRO Thioredoxin (TRX)... 27 7.0
TC88771 similar to GP|3643603|gb|AAC42250.1| unknown protein {Ar... 27 9.1
>TC87434 similar to SP|P52902|ODPA_PEA Pyruvate dehydrogenase E1 component
alpha subunit mitochondrial precursor (EC 1.2.4.1)
(PDHE1-A)., complete
Length = 1829
Score = 482 bits (1241), Expect = e-137
Identities = 235/288 (81%), Positives = 261/288 (90%)
Frame = +2
Query: 1 MALSRLASSSSSSSSTVGSKFLKPLTAAFSLRRSISSDSNPITIETSVPFTAHNCDPPSR 60
MALSRL SSSS+ST+ S +KP++ F+L RSISSD+ ++IETS+PFTAHNC PPS
Sbjct: 230 MALSRL---SSSSTSTIKSNLIKPISTIFTLNRSISSDTT-LSIETSIPFTAHNCTPPST 397
Query: 61 SVETSTAELHSFFRQMATMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITKKD 120
+V T+ EL +FF M+ MRRMEIAADSLYK+KLIRGFCHLYDGQEAVA+GMEA TKKD
Sbjct: 398 TVTTTPNELLNFFHTMSLMRRMEIAADSLYKSKLIRGFCHLYDGQEAVAVGMEASTTKKD 577
Query: 121 AIITAYRDHCTFLGRGGTLLEIFSELMGRVDGCSKGKGGSMHFYRKETNFYGGHGIVGAQ 180
IITAYRDHCTFLGRGGTLLE++SELMGRVDGCSKGKGGSMHFY+K++ F+GGHGIVGAQ
Sbjct: 578 CIITAYRDHCTFLGRGGTLLEVYSELMGRVDGCSKGKGGSMHFYKKDSGFFGGHGIVGAQ 757
Query: 181 VPLGCGVAFAQKYSKDEAVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMG 240
VPLGCG+AF QKY K+E+VTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMG
Sbjct: 758 VPLGCGLAFGQKYLKNESVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMG 937
Query: 241 TAEWRAAKSPAYYKRGDYVPGLKVDGMDVLAVKQACKFAKEHALKNGP 288
TAEWRAAKSPAYYKRGDYVPGLKVDGMDVLAVKQA KFAKEHAL+NGP
Sbjct: 938 TAEWRAAKSPAYYKRGDYVPGLKVDGMDVLAVKQAVKFAKEHALQNGP 1081
>TC86456 similar to SP|P52902|ODPA_PEA Pyruvate dehydrogenase E1 component
alpha subunit mitochondrial precursor (EC 1.2.4.1)
(PDHE1-A)., partial (92%)
Length = 1781
Score = 464 bits (1194), Expect = e-131
Identities = 229/290 (78%), Positives = 250/290 (85%), Gaps = 2/290 (0%)
Frame = +2
Query: 1 MALSRLASSSSSSSSTVGSKFLKPLTAAFSLR-RSISSDSNP-ITIETSVPFTAHNCDPP 58
MALSR SSS GS LKP + ++R RSISS S +T+ETS+PFT+HNC+PP
Sbjct: 203 MALSRF------SSSQFGSTLLKPYFLSSAIRHRSISSSSTETLTVETSIPFTSHNCEPP 364
Query: 59 SRSVETSTAELHSFFRQMATMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITK 118
S +V+T+ +EL SFF M MRRMEIAADSLYKAKLIRGFCHLYDGQEAVA+GMEA I +
Sbjct: 365 STTVQTTASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINR 544
Query: 119 KDAIITAYRDHCTFLGRGGTLLEIFSELMGRVDGCSKGKGGSMHFYRKETNFYGGHGIVG 178
KD +ITAYRDHCTFL RGGTL+E+FSELMGR DGCSKGKGGSMHFYRKE FYGGHGIVG
Sbjct: 545 KDCVITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVG 724
Query: 179 AQVPLGCGVAFAQKYSKDEAVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYG 238
AQ+PLG G+AF QKY+KD VTF LYGDGAANQGQLFEALNIAALWDLPAILVCENNHYG
Sbjct: 725 AQIPLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYG 904
Query: 239 MGTAEWRAAKSPAYYKRGDYVPGLKVDGMDVLAVKQACKFAKEHALKNGP 288
MGTAEWR+AKSPAYYKRGDY PGLKVDGMDVLAVKQACKFAKEHALKNGP
Sbjct: 905 MGTAEWRSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALKNGP 1054
>TC77795 similar to GP|15450707|gb|AAK96625.1 At1g01090/T25K16_8
{Arabidopsis thaliana}, partial (82%)
Length = 1690
Score = 180 bits (457), Expect = 5e-46
Identities = 106/285 (37%), Positives = 152/285 (53%), Gaps = 19/285 (6%)
Frame = +2
Query: 23 KPLTAAFSL-RRSISSDSNPITIETSVPFTAHNCDPPSRSV---ETSTAELHS-----FF 73
KPL+ + L + + SS S + + + A PP +V +TS + +
Sbjct: 257 KPLSFSSDLFKHNASSSSFLGSTQKLLRLNARRSPPPVAAVLLQQTSNLLITKEEGLVLY 436
Query: 74 RQMATMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITKKDAIITAYRDHCTFL 133
M R E +Y + GF HLY+GQEAV+ G + K+D I++ YRDH L
Sbjct: 437 EDMVLGRSFEDKCAEMYYRGKMFGFVHLYNGQEAVSTGFIKYLRKEDCIVSTYRDHVHAL 616
Query: 134 GRGGTLLEIFSELMGRVDGCSKGKGGSMHFYRKETNFYGGHGIVGAQVPLGCGVAFAQKY 193
+G E+ SEL G+ GC +G+GGSMH + KE N GG +G +P+ G AF+ KY
Sbjct: 617 SKGVPSREVMSELFGKATGCCRGQGGSMHMFSKEHNVLGGFAFIGEGIPVATGAAFSMKY 796
Query: 194 SKD-------EAVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRA 246
++ + VT A +GDG N GQ FE LN+AALW LP + V ENN + +G + RA
Sbjct: 797 KREVLNQADSDNVTLAFFGDGTCNNGQFFECLNMAALWKLPIVFVVENNLWAIGMSHLRA 976
Query: 247 AKSPAYYKRGDY--VPGLKVDGMDVLAVKQACKFAKEHALK-NGP 288
P +K+G +PG+ VDGMDVL V++ K A + A + GP
Sbjct: 977 TSDPEIWKKGPAFGMPGVHVDGMDVLKVREVAKEAIDRARRGEGP 1111
>TC78680 similar to GP|9454571|gb|AAF87894.1| branched-chain alpha keto-acid
dehydrogenase E1 - alpha subunit {Arabidopsis thaliana},
partial (76%)
Length = 1520
Score = 95.9 bits (237), Expect = 2e-20
Identities = 63/229 (27%), Positives = 103/229 (44%), Gaps = 5/229 (2%)
Frame = +3
Query: 62 VETSTAELHSFFRQMATMRRMEIAADSLYKAKLIRGFCHLY---DGQEAVAIGMEAGITK 118
V+ S + +M T++ M DS++ +G Y G+EAV I A ++
Sbjct: 354 VQVSKEMAVRMYSEMVTLQTM----DSIFYEVQRQGRISFYLTSMGEEAVNIASAAALSS 521
Query: 119 KDAIITAYRDHCTFLGRGGTLLEIFSELMGRVDGCSKGKGGSMHFYRKETNFYGGHGIVG 178
D I+ YR+ L RG TL + +L G + KG+ +H+ N++ +
Sbjct: 522 DDIILPQYREPGVLLWRGFTLQQFAHQLFGNTNDFGKGRQMPIHYGSNNHNYFTVSSPIA 701
Query: 179 AQVPLGCGVAFAQKYSKDEAVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYG 238
Q+P G A++ K A GDG ++G +N AA+ + P I +C NN +
Sbjct: 702 TQLPQAVGAAYSLKMDGKSACAVTFCGDGGTSEGDFHAGMNFAAVMEAPVIFICRNNGWA 881
Query: 239 MGTAEWRAAKSPAYYKRGDY--VPGLKVDGMDVLAVKQACKFAKEHALK 285
+ T +S +G + ++VDG D LAV A A+E A+K
Sbjct: 882 ISTPVEEQFRSDGIVVKGQAYGIWSIRVDGNDALAVYSAVHTAREIAIK 1028
>CA990749 similar to GP|9955517|emb branched-chain alpha keto-acid
dehydrogenase E1 alpha subunit-like protein {Arabidopsis
thaliana}, partial (53%)
Length = 799
Score = 94.0 bits (232), Expect = 6e-20
Identities = 53/184 (28%), Positives = 87/184 (46%), Gaps = 2/184 (1%)
Frame = +2
Query: 103 DGQEAVAIGMEAGITKKDAIITAYRDHCTFLGRGGTLLEIFSELMGRVDGCSKGKGGSMH 162
+G+EA+ I A ++ D I YR+ L RG TL E ++ KG+ H
Sbjct: 17 NGEEAINIASAAALSMNDVIFPQYREQGVLLWRGFTLQEFANQCFSNKFDNGKGRQMPAH 196
Query: 163 FYRKETNFYGGHGIVGAQVPLGCGVAFAQKYSKDEAVTFALYGDGAANQGQLFEALNIAA 222
+ + N+ V Q+P G A++ K K +A +GDG +++G LN AA
Sbjct: 197 YGSNKHNYMNVASTVATQIPHAVGAAYSLKMDKKDACAVTYFGDGGSSEGDFHAGLNFAA 376
Query: 223 LWDLPAILVCENNHYGMGTAEWRAAKSPAYYKRGDY--VPGLKVDGMDVLAVKQACKFAK 280
+ + P I +C NN + + T +S +G V ++VDG D LA+ A + A+
Sbjct: 377 VMEAPVIFICRNNGWAISTPTSDQFRSDGIVVKGQAYGVRSIRVDGNDALAIYSAVQAAR 556
Query: 281 EHAL 284
+ A+
Sbjct: 557 QMAV 568
>TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Ateline
herpesvirus 3}, partial (8%)
Length = 986
Score = 30.8 bits (68), Expect = 0.63
Identities = 23/59 (38%), Positives = 32/59 (53%), Gaps = 3/59 (5%)
Frame = -2
Query: 4 SRLASSSSSSSSTVGSKFLKPLTAAFSLRRSISSD---SNPITIETSVPFTAHNCDPPS 59
S +SSS SSSST S P +++ S S SS S+P + +S PF++ PPS
Sbjct: 736 SSSSSSSLSSSSTPSSSSSSPPSSSSSSSSSSSSSSSWSSPSSSSSSSPFSSAPPAPPS 560
>TC89165 similar to PIR|S19652|S19652 cellodextrinase C - Pseudomonas
fluorescens, partial (2%)
Length = 1018
Score = 30.4 bits (67), Expect = 0.82
Identities = 17/34 (50%), Positives = 21/34 (61%)
Frame = +3
Query: 3 LSRLASSSSSSSSTVGSKFLKPLTAAFSLRRSIS 36
L + S SSSSSS+ S F + AFS+RRS S
Sbjct: 375 LGKSCSVSSSSSSSTASGFQTTMKRAFSIRRSSS 476
>TC85485 similar to GP|22136900|gb|AAM91794.1 putative transketolase
{Arabidopsis thaliana}, partial (90%)
Length = 2728
Score = 29.6 bits (65), Expect = 1.4
Identities = 20/73 (27%), Positives = 37/73 (50%), Gaps = 11/73 (15%)
Frame = +2
Query: 175 GIVGAQVPLGCGVAFAQK-----YSKDEA-----VTFALYGDGAANQGQLFEALNIAALW 224
G +G + G G+A A+K ++K ++ T+ + GDG +G EA ++A W
Sbjct: 746 GPLGQGIANGVGLALAEKHLAARFNKPDSEIVDHYTYVILGDGCQMEGISNEACSLAGHW 925
Query: 225 DLPAILV-CENNH 236
L ++ ++NH
Sbjct: 926 GLGKLIAFYDDNH 964
>TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed protein
{Arabidopsis thaliana}, partial (50%)
Length = 753
Score = 28.9 bits (63), Expect = 2.4
Identities = 26/73 (35%), Positives = 37/73 (50%), Gaps = 3/73 (4%)
Frame = -2
Query: 4 SRLASSSSSSSSTVGSKFLKPLTAAFSLRRSISSDSNP---ITIETSVPFTAHNCDPPSR 60
S L+SSSSSSSS+ S P S + SS S+P ++ +S FT+ + S
Sbjct: 494 SSLSSSSSSSSSSSSSLPPAPFGTGSSSGSASSSSSSPEKSSSLSSSSSFTSSS----SS 327
Query: 61 SVETSTAELHSFF 73
S +S+ L S F
Sbjct: 326 SSSSSSVSLPSLF 288
>BM778905 similar to SP|Q05046|CH62_ Chaperonin CPN60-2 mitochondrial
precursor (HSP60-2). [Pumpkin Winter squash] {Cucurbita
maxima}, partial (9%)
Length = 456
Score = 28.5 bits (62), Expect = 3.1
Identities = 17/28 (60%), Positives = 21/28 (74%), Gaps = 2/28 (7%)
Frame = +1
Query: 6 LASSSSSSSSTVG--SKFLKPLTAAFSL 31
L+SSSSSSSST+G +F KPL A +L
Sbjct: 328 LSSSSSSSSSTLGFVERFSKPLPAIRNL 411
>TC80072 similar to GP|14423520|gb|AAK62442.1 Unknown protein {Arabidopsis
thaliana}, partial (17%)
Length = 746
Score = 28.1 bits (61), Expect = 4.1
Identities = 16/34 (47%), Positives = 23/34 (67%)
Frame = -1
Query: 7 ASSSSSSSSTVGSKFLKPLTAAFSLRRSISSDSN 40
+SSSSSSSS+ S + L+ + SL+ ISS S+
Sbjct: 539 SSSSSSSSSSSPSSSVSSLSISLSLKIGISSSSS 438
>TC87823 similar to GP|9295275|gb|AAF86907.1| phosphoenolpyruvate/phosphate
translocator precursor {Mesembryanthemum crystallinum},
partial (74%)
Length = 1552
Score = 28.1 bits (61), Expect = 4.1
Identities = 27/71 (38%), Positives = 38/71 (53%), Gaps = 3/71 (4%)
Frame = +3
Query: 6 LASSSSSSSSTVGSKFLKPLTAAFSLR---RSISSDSNPITIETSVPFTAHNCDPPSRSV 62
L+SSSSSSS T S + P +++F R +SD +P TSVP +A + S S+
Sbjct: 207 LSSSSSSSSFTRQSWSMSPSSSSFKFRPLPLVPTSDLSP-PQATSVPESAGDSSAESSSL 383
Query: 63 ETSTAELHSFF 73
T +L S F
Sbjct: 384 -LKTLQLGSLF 413
>BQ751067 similar to GP|18461169|dbj hypothetical protein~similar to
Arabidopsis thaliana chromosome 5 MCL19.15, partial
(3%)
Length = 754
Score = 27.7 bits (60), Expect = 5.3
Identities = 16/48 (33%), Positives = 27/48 (55%), Gaps = 1/48 (2%)
Frame = +3
Query: 22 LKPLTAAFSLRRS-ISSDSNPITIETSVPFTAHNCDPPSRSVETSTAE 68
LKP+ A S +S+ ++P T+ P + H C+PP +S +T+E
Sbjct: 144 LKPMADASRPHMS*LSASAHP----TNPPRS*HQCNPPKQSTRATTSE 275
>TC85484 similar to PIR|S58083|S58083 transketolase (EC 2.2.1.1) precursor -
potato (fragment), partial (95%)
Length = 2685
Score = 27.3 bits (59), Expect = 7.0
Identities = 19/73 (26%), Positives = 34/73 (46%), Gaps = 11/73 (15%)
Frame = +1
Query: 175 GIVGAQVPLGCGVAFAQKY------SKDEAV----TFALYGDGAANQGQLFEALNIAALW 224
G +G + G+A A+K+ D + T+ + GDG +G EA ++A W
Sbjct: 652 GPLGQGIANAVGLALAEKHLAARFNKPDNEIIDHYTYCILGDGCQMEGIANEACSLAGHW 831
Query: 225 DLPAILV-CENNH 236
L ++ ++NH
Sbjct: 832 GLGKLIAFYDDNH 870
>TC77049 homologue to GP|3941289|gb|AAC82326.1| similarity to SCAMP37 {Pisum
sativum}, complete
Length = 1358
Score = 27.3 bits (59), Expect = 7.0
Identities = 16/42 (38%), Positives = 22/42 (52%)
Frame = +1
Query: 10 SSSSSSTVGSKFLKPLTAAFSLRRSISSDSNPITIETSVPFT 51
SS +++TV + PL A F L + DSNP E PF+
Sbjct: 166 SSLNNTTVATFLYSPLLACFYLIMAGRYDSNPFDEEQVNPFS 291
>TC83037 weakly similar to PIR|T38758|T38758 60s ribosomal protein L5 -
fission yeast (Schizosaccharomyces pombe), partial (49%)
Length = 844
Score = 27.3 bits (59), Expect = 7.0
Identities = 18/40 (45%), Positives = 26/40 (65%), Gaps = 2/40 (5%)
Frame = -2
Query: 5 RLASSSSSSSSTVGSKFLKP--LTAAFSLRRSISSDSNPI 42
R ASSSSS +T+ S + P L+A SL R++ + SNP+
Sbjct: 126 RGASSSSSMGATLPSGSVMP*YLSANSSLVRTLRAASNPV 7
>TC87825 similar to GP|21553995|gb|AAM63076.1 PP2A regulatory subunit
{Arabidopsis thaliana}, partial (94%)
Length = 1448
Score = 27.3 bits (59), Expect = 7.0
Identities = 15/22 (68%), Positives = 17/22 (77%)
Frame = -3
Query: 1 MALSRLASSSSSSSSTVGSKFL 22
+ALS ASSSSSSSS+ G FL
Sbjct: 1200 LALSCAASSSSSSSSSPGLNFL 1135
>TC78799 weakly similar to SP|P96132|THIO_THIRO Thioredoxin (TRX)
(Fragment). {Thiocapsa roseopersicina}, partial (37%)
Length = 734
Score = 27.3 bits (59), Expect = 7.0
Identities = 9/15 (60%), Positives = 13/15 (86%)
Frame = -2
Query: 225 DLPAILVCENNHYGM 239
DL +I++C+ NHYGM
Sbjct: 352 DLTSIIICQLNHYGM 308
>TC88771 similar to GP|3643603|gb|AAC42250.1| unknown protein {Arabidopsis
thaliana}, partial (33%)
Length = 1295
Score = 26.9 bits (58), Expect = 9.1
Identities = 17/56 (30%), Positives = 28/56 (49%)
Frame = +1
Query: 10 SSSSSSTVGSKFLKPLTAAFSLRRSISSDSNPITIETSVPFTAHNCDPPSRSVETS 65
SSSSSS++ ++ + L+ S RR ++ S P P TA + P + T+
Sbjct: 70 SSSSSSSMAAQLFRDLSLGHSKRRDSTTPSPPSLKIMPPPPTADDLPSPLGQLSTN 237
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.134 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,758,534
Number of Sequences: 36976
Number of extensions: 120205
Number of successful extensions: 900
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 774
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 847
length of query: 288
length of database: 9,014,727
effective HSP length: 95
effective length of query: 193
effective length of database: 5,502,007
effective search space: 1061887351
effective search space used: 1061887351
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0028b.3


