

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0028b.2
(533 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
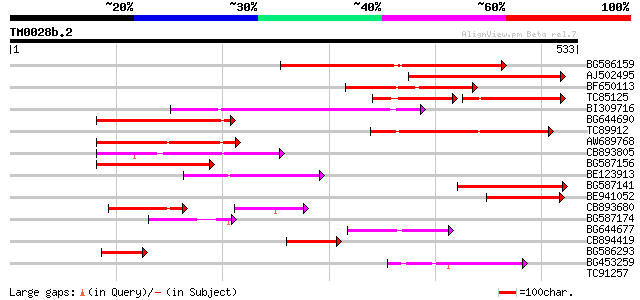
Score E
Sequences producing significant alignments: (bits) Value
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 204 9e-53
AJ502495 weakly similar to GP|18071369|g putative gag-pol polypr... 160 1e-39
BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vu... 139 3e-33
TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-relate... 85 1e-32
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 134 9e-32
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 132 4e-31
TC89912 weakly similar to PIR|B84512|B84512 probable retroelemen... 124 1e-28
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 119 4e-27
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 115 3e-26
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 104 7e-23
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 83 3e-16
BG587141 similar to PIR|H86461|H86 hypothetical protein AAF32440... 81 1e-15
BE941052 weakly similar to PIR|B85188|B85 retrotransposon like p... 66 4e-11
CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotia... 64 1e-10
BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse ... 63 2e-10
BG644677 weakly similar to GP|7682800|gb| Hypothetical protein T... 57 2e-08
CB894419 weakly similar to GP|10177935|db copia-type polyprotein... 52 7e-07
BG586293 weakly similar to PIR|E84473|E84 probable retroelement ... 50 2e-06
BG453259 homologue to GP|21434|emb|CA ORF4 {Solanum tuberosum}, ... 49 6e-06
TC91257 similar to GP|21434|emb|CAA36616.1|| ORF4 {Solanum tuber... 37 0.025
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 204 bits (518), Expect = 9e-53
Identities = 102/214 (47%), Positives = 145/214 (67%), Gaps = 1/214 (0%)
Frame = +1
Query: 255 GIFMYQQKYIKDILKRFNMGNCHPVTTPSEVNIKLRRDDEG-DVDASLYKQLVGSLRFLC 313
GI++ Q+KY+ D+L+RF M + P KL +D+ G VDA+ YKQ+VG L +L
Sbjct: 25 GIYICQRKYVTDLLERFGMEKSNLSRNPIAPRCKLIKDENGVKVDATKYKQIVGCLMYLA 204
Query: 314 NSRLDISYGVGLISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKLGKELELVGY 373
+R D+ Y + LIS+FM P + H+ A KR+LRYL GT + GI++ R G E +L Y
Sbjct: 205 ATRPDLMYVLSLISRFMNCPTELHMHAVKRVLRYLNGTINLGIMYKRN---GSE-KLEAY 372
Query: 374 TDSDYSGDLDDRKSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLE 433
TDSDY+GDLDDRKSTSGY+F L +SW+SKKQPV+ LS+ +AE+IA +F ACQ+VW+
Sbjct: 373 TDSDYAGDLDDRKSTSGYVFMLSSGAVSWSSKKQPVVTLSTTKAEFIAAAFCACQSVWMR 552
Query: 434 ELLKEMGFVTKKPMELMVDNISAISLAKNPISHG 467
+L+++G+ + + DN S I L+KNP+ HG
Sbjct: 553 RVLEKLGYTQSGSITMYCDNNSTIKLSKNPVLHG 654
>AJ502495 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (9%)
Length = 542
Score = 160 bits (405), Expect = 1e-39
Identities = 77/147 (52%), Positives = 105/147 (71%)
Frame = +2
Query: 376 SDYSGDLDDRKSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLEEL 435
SD++GD + RKSTSGY F L ISW+SKKQPV+A S+ EAEYIA + A Q VWL +
Sbjct: 2 SDWAGDTETRKSTSGYAFHLGTGAISWSSKKQPVVAFSTAEAEYIASTSCATQTVWLRRI 181
Query: 436 LKEMGFVTKKPMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKLELVYCPT 495
L+ M P ++ DN SAI+L+KNP+ HGRSKHI+++FH +RE + ++ + YCPT
Sbjct: 182 LEVMHHEQNTPTKIYCDNKSAIALSKNPVFHGRSKHIDIQFHKIRELIAEKEVVIEYCPT 361
Query: 496 DEQKADIFTKPLKTERFVILRKEIGVV 522
+E+ ADIFTKPLK E F L+K +G++
Sbjct: 362 EEKIADIFTKPLKIESFYKLKKMLGMM 442
>BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vulgaris},
partial (13%)
Length = 494
Score = 139 bits (350), Expect = 3e-33
Identities = 71/124 (57%), Positives = 93/124 (74%)
Frame = +1
Query: 316 RLDISYGVGLISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKLGKELELVGYTD 375
R DI Y V +ISKFM P+K HL AA RILRY++GT +YG+LFP K + EL+ Y+D
Sbjct: 121 RPDICYSVSVISKFMHDPRKPHLIAANRILRYVRGTMEYGLLFPYGAK-SEVYELICYSD 297
Query: 376 SDYSGDLDDRKSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLEEL 435
SD+ GD R+STSGY+F+ + A ISW +KKQP+ ALSS EAEYIAG+FA QA+WL+ +
Sbjct: 298 SDWCGD---RRSTSGYVFKFNDAAISWCTKKQPITALSSYEAEYIAGTFATFQALWLDSV 468
Query: 436 LKEM 439
+KE+
Sbjct: 469 IKEL 480
>TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
(EC 3.4.23.-);, partial (7%)
Length = 705
Score = 85.1 bits (209), Expect(2) = 1e-32
Identities = 42/80 (52%), Positives = 56/80 (69%)
Frame = +1
Query: 342 KRILRYLQGTTDYGILFPRQKKLGKELELVGYTDSDYSGDLDDRKSTSGYLFQLHGAPIS 401
KRI+RY++GT+ + F G EL + GY DSD++GD D RKST+GY+F L G +S
Sbjct: 4 KRIMRYIKGTSGVAVCFG-----GSELTVRGYVDSDFAGDHDKRKSTTGYVFTLAGGAVS 168
Query: 402 WTSKKQPVIALSSCEAEYIA 421
W SK Q V+ALS+ EAEY+A
Sbjct: 169 WLSKLQTVVALSTTEAEYMA 228
Score = 73.6 bits (179), Expect(2) = 1e-32
Identities = 33/98 (33%), Positives = 64/98 (64%), Gaps = 1/98 (1%)
Frame = +3
Query: 426 AC-QAVWLEELLKEMGFVTKKPMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVN 484
AC +A+W++ L++E+G ++ + + D+ SA+ +A+NP H R+KHI +++HF+RE V
Sbjct: 240 ACKEAIWMQRLMEELGH-KQEQITVYCDSQSALHIARNPAFHSRTKHIGIQYHFVREVVE 416
Query: 485 GGKLELVYCPTDEQKADIFTKPLKTERFVILRKEIGVV 522
G +++ T++ AD TK + T++F+ R G++
Sbjct: 417 EGSVDMQKIHTNDNLADAMTKSINTDKFIWCRSSYGLL 530
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 134 bits (337), Expect = 9e-32
Identities = 75/241 (31%), Positives = 133/241 (55%), Gaps = 1/241 (0%)
Frame = +2
Query: 152 VFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHGDTGLLLICLYVDD 211
V +L K++YGLKQA R W ++ L+ + + S ++ ++ + D+ + +YVDD
Sbjct: 20 VCELQKSIYGLKQASRQWYSKLSESLISFGYLQSSSDFSLFTKFK--DSSFTTLLVYVDD 193
Query: 212 LLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQFEYTAAGIFMYQQKYIKDILKRF 271
+++ G+D E+ +K L F++ DLG L YFLG++ + GI + Q+KY ++L+
Sbjct: 194 IVLAGNDISEIQHVKCFLIDRFKIKDLGSLRYFLGLEVARSKQGILLNQRKYTLELLEDS 373
Query: 272 NMGNCHPVTTPSEVNIKLRRDDEGDV-DASLYKQLVGSLRFLCNSRLDISYGVGLISKFM 330
TP ++++KL D D + Y++L+G L +L +R DIS+ V +S+F+
Sbjct: 374 GNLAVKSTLTPYDISLKLHNSDSPLYNDETQYRRLIGKLIYLTTTRPDISFAVQQLSQFV 553
Query: 331 CAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKLGKELELVGYTDSDYSGDLDDRKSTSG 390
P++ H AA R+L+YL+ G+ + L+L + DSD++ RKS +G
Sbjct: 554 SKPQQVHYQAAIRVLQYLKTAPAKGLFY----SATSNLKLSSFADSDWATCPTTRKSVTG 721
Query: 391 Y 391
Y
Sbjct: 722 Y 724
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 132 bits (331), Expect = 4e-31
Identities = 64/131 (48%), Positives = 95/131 (71%)
Frame = -2
Query: 82 FSTKPGLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPP 141
++ K G+DY E F+PVAR+E IR++++ A+ + ++Q+DVKSAF+NG L+EEV+V QPP
Sbjct: 388 YNQKEGIDYDEAFSPVARMEAIRILIAFAAFMGFKLYQMDVKSAFINGDLKEEVFVKQPP 209
Query: 142 GFEIQGKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHGDTG 201
GFE N VF+L+K LYGLKQAPRAW +R+ FLL+ F R ++ +++ K +
Sbjct: 208 GFEDAEVPNHVFRLNKTLYGLKQAPRAWYERLSKFLLKNGFKRGKIDNTLFLLKRE*E-- 35
Query: 202 LLLICLYVDDL 212
LL+I +YVDD+
Sbjct: 34 LLIIQVYVDDI 2
>TC89912 weakly similar to PIR|B84512|B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(10%)
Length = 814
Score = 124 bits (310), Expect = 1e-28
Identities = 63/172 (36%), Positives = 109/172 (62%)
Frame = +1
Query: 340 AAKRILRYLQGTTDYGILFPRQKKLGKELELVGYTDSDYSGDLDDRKSTSGYLFQLHGAP 399
A K +L+YL + + + + + +E L GY D+DY+G++D RKS SG++F L+G
Sbjct: 4 ALKWVLKYLNESLKSSLKYTKAAQ--EEDALEGYVDADYAGNVDTRKSLSGFVFTLYGTT 177
Query: 400 ISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLEELLKEMGFVTKKPMELMVDNISAISL 459
ISW + +Q V+ LS+ +AEYIA A+WL+ ++ E+G +T++ +++ D+ SAI L
Sbjct: 178 ISWKANQQSVVTLSTTQAEYIAFVEGVKDAIWLKGMIGELG-ITQEYVKIHCDSQSAIHL 354
Query: 460 AKNPISHGRSKHIEVRFHFLREQVNGGKLELVYCPTDEQKADIFTKPLKTER 511
A + + H R+KHI++R HF+R+ + ++ + ++E AD+FTK K R
Sbjct: 355 ANHQVYHERTKHIDIRLHFIRDMIESKEIVVEKMASEENPADVFTKLKKHAR 510
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 119 bits (297), Expect = 4e-27
Identities = 63/136 (46%), Positives = 89/136 (65%)
Frame = +1
Query: 82 FSTKPGLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPP 141
FS G DY+E F+PV + TIRL+++IA W I Q+D+ +AFLNG L+EEVY+SQP
Sbjct: 268 FSQTLGCDYTETFSPVIKPVTIRLILTIAITYKWEIQQIDINNAFLNGFLQEEVYMSQPQ 447
Query: 142 GFEIQGKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHGDTG 201
GFE K + V KL+K+LYGLKQAPRAW + + +Q F + + + + +G
Sbjct: 448 GFEAANK-SLVCKLNKSLYGLKQAPRAWYEXLTSAQIQFGFTKSRCDPSLLIYNQNG--A 618
Query: 202 LLLICLYVDDLLITGS 217
+ + +YVDD+LITGS
Sbjct: 619 CIYLXIYVDDILITGS 666
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 115 bits (289), Expect = 3e-26
Identities = 65/180 (36%), Positives = 108/180 (59%), Gaps = 3/180 (1%)
Frame = +3
Query: 82 FSTKPGLDYSEVFAPVARIETIRLVVSIASARNWP---IFQLDVKSAFLNGPLEEEVYVS 138
+S + G+DY+EVFAPVAR +TIR+V+++A+ I + + + + + + ++
Sbjct: 249 YSQQYGVDYTEVFAPVARWDTIRMVIALAAQIKRDGVCIS*M*KAHSCMEN*MRKFLLIN 428
Query: 139 QPPGFEIQGKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHG 198
+ + ++ +ALYGLKQAPRAW RI+ + + F +C E+ ++V+ S G
Sbjct: 429 H----RVM*RRVIS*RVKRALYGLKQAPRAWYSRIEAYFTKEGFEKCPYEHTLFVKLSEG 596
Query: 199 DTGLLLICLYVDDLLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQFEYTAAGIFM 258
+L+I LYVDDL+ G+D +E K+ +K EF MSDLGK+ YFLG++ GI++
Sbjct: 597 GK-ILIISLYVDDLIFIGNDENMFEEFKKSMKKEFNMSDLGKMHYFLGVEVTQNEKGIYI 773
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 104 bits (260), Expect = 7e-23
Identities = 50/111 (45%), Positives = 74/111 (66%)
Frame = -1
Query: 82 FSTKPGLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPP 141
F+ G DY E FAPVA++ TIR+V+S+A W ++Q+DVK+AFL G LE+EVY+ PP
Sbjct: 351 FTLTYGEDYIETFAPVAKLHTIRIVLSLAVNLGWGLWQMDVKNAFLQGELEDEVYMYPPP 172
Query: 142 GFEIQGKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVY 192
G E K +V +L KA+YGLKQ+PRAW ++ L F + +++ ++
Sbjct: 171 GLEHLVKRGNVLRLKKAIYGLKQSPRAWYNKLSTTLNGRGFRKSELDHTLF 19
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 82.8 bits (203), Expect = 3e-16
Identities = 44/133 (33%), Positives = 70/133 (52%)
Frame = +1
Query: 164 QAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHGDTGLLLICLYVDDLLITGSDPKELD 223
Q+PR W R + + + +C ++ ++++ S +LI +YVDD+ +TG K +
Sbjct: 1 QSPRDWFDRFT*VVKKFGYIQCQTDHAMFIKHSSTVKKAILI-VYVDDIFLTGDHGK*IK 177
Query: 224 ELKQILKSEFEMSDLGKLGYFLGMQFEYTAAGIFMYQQKYIKDILKRFNMGNCHPVTTPS 283
LK +L EFE+ DLG L YFLGM+ G + Q+KY+ D+LK M C + P
Sbjct: 178 RLKNLLAEEFEIKDLGNLKYFLGMEVARWKKGSSISQRKYVLDLLKETRMIGCKTIRDPY 357
Query: 284 EVNIKLRRDDEGD 296
N + R + D
Sbjct: 358 GCNCEARNSRQWD 396
Score = 40.4 bits (93), Expect = 0.002
Identities = 23/54 (42%), Positives = 33/54 (60%), Gaps = 1/54 (1%)
Frame = +2
Query: 278 PVTTPSEVNIKLRRDDEGD-VDASLYKQLVGSLRFLCNSRLDISYGVGLISKFM 330
P TP + +KL D G VD Y++LVG L +L ++R DIS+ V +S+FM
Sbjct: 341 PSETPMDATVKLGTLDNGTLVDKGRYQRLVGKLIYLSHTRPDISFVVCTMSQFM 502
>BG587141 similar to PIR|H86461|H86 hypothetical protein AAF32440.1
[imported] - Arabidopsis thaliana, partial (20%)
Length = 731
Score = 80.9 bits (198), Expect = 1e-15
Identities = 41/103 (39%), Positives = 65/103 (62%)
Frame = +3
Query: 422 GSFAACQAVWLEELLKEMGFVTKKPMELMVDNISAISLAKNPISHGRSKHIEVRFHFLRE 481
G AA QA+WL++LL E+ + + + + +DN S I+L +NP+ HGR HI R+HF+RE
Sbjct: 117 GPTAARQAMWLQDLLSEVTWEPCEEVVIRIDNQSVIALTRNPVFHGRGNHIHKRYHFIRE 296
Query: 482 QVNGGKLELVYCPTDEQKADIFTKPLKTERFVILRKEIGVVSL 524
V G++E+ + P ++ +A I TK L F +R IG++ L
Sbjct: 297 CVENGQVEVEHVPGEKHRAYI*TKALGRIIFREIRYYIGMIDL 425
>BE941052 weakly similar to PIR|B85188|B85 retrotransposon like protein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 480
Score = 65.9 bits (159), Expect = 4e-11
Identities = 33/73 (45%), Positives = 44/73 (60%)
Frame = +2
Query: 449 LMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKLELVYCPTDEQKADIFTKPLK 508
L D +SA L NP+ H R KHI + HF+R+ V GKL++ + T +Q AD TKPL
Sbjct: 23 LRCDYLSATYLTHNPVYHSRMKHISIDIHFVRDLVQQGKLKVQHVCTVDQLADCLTKPLS 202
Query: 509 TERFVILRKEIGV 521
R +LR +IGV
Sbjct: 203 KSRHQLLRNKIGV 241
>CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotiana tabacum},
partial (7%)
Length = 780
Score = 64.3 bits (155), Expect = 1e-10
Identities = 35/75 (46%), Positives = 49/75 (64%), Gaps = 1/75 (1%)
Frame = -2
Query: 94 FAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPPGFEIQ-GKENSV 152
F P+ ++ TI ++SI + N + LDVK+AFL G L E++Y+ QP GF + GK V
Sbjct: 554 FVPIVKLNTIMFLLSIVAIENLYLE*LDVKTAFLRGDLVEDIYMHQPEGFS*EVGK--MV 381
Query: 153 FKLSKALYGLKQAPR 167
KL K++YGLKQ PR
Sbjct: 380 GKLKKSMYGLKQGPR 336
Score = 42.7 bits (99), Expect = 3e-04
Identities = 26/72 (36%), Positives = 38/72 (52%), Gaps = 2/72 (2%)
Frame = -3
Query: 212 LLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQ--FEYTAAGIFMYQQKYIKDILK 269
LL+ GS+ E+ LK E +M DLG +GMQ + + + Q +YI +L+
Sbjct: 271 LLVVGSNIDEIKNLKTRFSKEIDMKDLGPAKKIIGMQIMIDKQKGVL*LSQVEYITRVLQ 92
Query: 270 RFNMGNCHPVTT 281
FNMGN V+T
Sbjct: 91 IFNMGNAILVST 56
>BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse
transcriptase homolog - rape retrotransposon copia-like
(fragment), partial (84%)
Length = 249
Score = 63.2 bits (152), Expect = 2e-10
Identities = 38/99 (38%), Positives = 50/99 (50%), Gaps = 16/99 (16%)
Frame = -1
Query: 131 LEEEVYVSQPPGFEIQGKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYG 190
LEE++Y++QP GF GKE+ V KL K+LYGLKQ+PR W KR D +
Sbjct: 246 LEEKIYMTQPEGFLFPGKEDHVCKLRKSLYGLKQSPRQWYKRFDSY-------------- 109
Query: 191 VYVRKSHGDTGLLL----------------ICLYVDDLL 213
R S TG+L+ + LYVDD+L
Sbjct: 108 ---RSSWATTGVLMTVVST*TR*RMSRYIYLVLYVDDML 1
>BG644677 weakly similar to GP|7682800|gb| Hypothetical protein T15F17.l
{Arabidopsis thaliana}, partial (3%)
Length = 539
Score = 57.0 bits (136), Expect = 2e-08
Identities = 32/100 (32%), Positives = 51/100 (51%)
Frame = -3
Query: 318 DISYGVGLISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKLGKELELVGYTDSD 377
+I++ + L+S++ AP H K I +YL+G D G+ + + +L+GY ++
Sbjct: 507 NITFSINLLSRYSSAPTMRH*NGIKHICKYLKGIIDMGLFYSKDCSP----DLIGYVNA* 340
Query: 378 YSGDLDDRKSTSGYLFQLHGAPISWTSKKQPVIALSSCEA 417
Y D +S +GY+F ISW S K IA SS A
Sbjct: 339 YLSDPHKARS*TGYIFTCGNTVISWRSTK*STIATSSNHA 220
>CB894419 weakly similar to GP|10177935|db copia-type polyprotein
{Arabidopsis thaliana}, partial (2%)
Length = 170
Score = 51.6 bits (122), Expect = 7e-07
Identities = 25/53 (47%), Positives = 37/53 (69%), Gaps = 1/53 (1%)
Frame = +3
Query: 261 QKYIKDILKRFNMGNCHPVTTPSEVNIKLRRDDEGD-VDASLYKQLVGSLRFL 312
+K DILK+F M + P++TP E +KL R+ +G VD++ YK L+GSLR+L
Sbjct: 3 KKCASDILKKFKMEHSKPISTPVEEKLKLTRESDGKRVDSTHYKSLIGSLRYL 161
>BG586293 weakly similar to PIR|E84473|E84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 763
Score = 50.1 bits (118), Expect = 2e-06
Identities = 24/43 (55%), Positives = 32/43 (73%)
Frame = +2
Query: 87 GLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNG 129
G+D+ EVFAPV RIETI L++++A+ I +DVK AFLNG
Sbjct: 194 GIDFDEVFAPVVRIETI*LLLALAATNGC*IHHIDVKIAFLNG 322
>BG453259 homologue to GP|21434|emb|CA ORF4 {Solanum tuberosum}, partial (5%)
Length = 657
Score = 48.5 bits (114), Expect = 6e-06
Identities = 42/133 (31%), Positives = 64/133 (47%), Gaps = 2/133 (1%)
Frame = -2
Query: 356 ILFPRQKKLGKELELVGYTDSDYSGDLDDRKSTSGYLFQLHGAPISWTSKKQPVIA--LS 413
I+F R+ KL E+ Y ++ +G + DR STSGY L G + KQ V+A +
Sbjct: 530 IVFKRE*KLSMEV----YXNTVCAGWIVDRGSTSGY*MFLGG---NMVE*KQNVVAR*VQ 372
Query: 414 SCEAEYIAGSFAACQAVWLEELLKEMGFVTKKPMELMVDNISAISLAKNPISHGRSKHIE 473
E + L+ L ++ K PM L +N +A NP+ H R+KHIE
Sbjct: 371 RHNFELCSQGL*RVMDEELKIKLDDLIINYKDPMTLF*NNNFVSRIAHNPVQHYRTKHIE 192
Query: 474 VRFHFLREQVNGG 486
+ HF+ E++ G
Sbjct: 191 IDQHFIIEKLYSG 153
>TC91257 similar to GP|21434|emb|CAA36616.1|| ORF4 {Solanum tuberosum},
partial (8%)
Length = 854
Score = 36.6 bits (83), Expect = 0.025
Identities = 17/40 (42%), Positives = 27/40 (67%), Gaps = 2/40 (5%)
Frame = +3
Query: 293 DEGDV--DASLYKQLVGSLRFLCNSRLDISYGVGLISKFM 330
D+G+ D Y++LVG L +L +R DISY V ++S+F+
Sbjct: 729 DQGETFSDPGRYRRLVGKLNYLTMTRPDISYAVSVVSQFL 848
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.331 0.146 0.454
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,911,040
Number of Sequences: 36976
Number of extensions: 212959
Number of successful extensions: 1026
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 1003
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1012
length of query: 533
length of database: 9,014,727
effective HSP length: 101
effective length of query: 432
effective length of database: 5,280,151
effective search space: 2281025232
effective search space used: 2281025232
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0028b.2


