

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0026.5
(1710 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
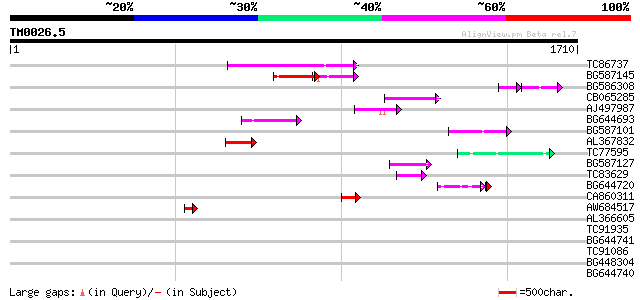
Score E
Sequences producing significant alignments: (bits) Value
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 192 7e-49
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 135 1e-31
BG586308 weakly similar to PIR|F84528|F8 probable retroelement p... 62 9e-22
CB065285 weakly similar to PIR|A84500|A84 probable retroelement ... 97 7e-20
AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis... 91 3e-18
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 90 7e-18
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 87 7e-17
AL367832 weakly similar to GP|14091845|gb Putative retroelement ... 86 9e-17
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 77 6e-14
BG587127 weakly similar to PIR|H84506|H84 probable retroelement ... 65 2e-10
TC83629 weakly similar to GP|6692261|gb|AAF24611.1| putative RNa... 53 9e-07
BG644720 45 2e-06
CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product ... 47 5e-05
AW684517 similar to GP|9927273|dbj Similar to Arabidopsis thalia... 44 5e-04
AL366605 41 0.003
TC91935 41 0.005
BG644741 40 0.008
TC91086 39 0.023
BG448304 37 0.086
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 36 0.11
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 192 bits (489), Expect = 7e-49
Identities = 133/403 (33%), Positives = 211/403 (52%), Gaps = 9/403 (2%)
Frame = +1
Query: 658 LLKEFKDCFAWDYDEMPGLSRDLVELKLPIKEDKK------PVKQLPRRFHPDVLVKIKE 711
+L+EF D F + SR L++ +P+ DK P L ++LV +K+
Sbjct: 343 VLEEFPDLFNPEKAYQVPASRGLLDHAIPLIPDKDGNDPPLPWGPLYGMSRQELLV-LKK 519
Query: 712 EIERLLKCKFIRTARYVDWLANVVPVIKKNGKMRVCIDFRDLNAATPKDEYHMPVAEMMV 771
+E LL FI+ + A V+ V K G +R C+D+R LNA T KD Y +P+ +
Sbjct: 520 TLEDLLDKGFIKASGSAAG-APVLFVRKPGGGIRFCVDYRALNAITKKDRYPLPLISETL 696
Query: 772 DSTAGHEYLSLLDGYSGYNQIFIAEEDVSKTAFRCPGALGTYEWVVMPFGLKNAGATYQR 831
AG + + LD + ++++ I +ED KTAFR G +EW+V PFGL A AT+QR
Sbjct: 697 RRVAGARWFTKLDVVAAFHKMRIKDEDQEKTAFRT--RYGLFEWIVCPFGLTGAPATFQR 870
Query: 832 VMNTIFHDFIETFMQVYIDDIVV-KSPSRDDHLAHLRKSFERMRKYGLKMNPLKCAFGVI 890
+N H+F++ F+ YIDD+++ + S+ DH A +R+ R+ GL ++P KC F V
Sbjct: 871 YINKTLHEFLDDFVTAYIDDVLIYTTGSKKDHEAQVRRVLRRLADAGLSLDPKKCEFSVT 1050
Query: 891 AGDFLGFVVHK-KGIEINKNKAKAILDTSPPTSKKQLQSLLGKINFLRRFIANLSEKTKS 949
++GF++ KG+ + K AI D PP S K +S LG N+ + FI SE T+
Sbjct: 1051 TVKYVGFILTAGKGVSCDPLKLAAIRDWLPPGSVKGARSFLGFCNYYKDFIPGYSEITE- 1227
Query: 950 FSPLLRLKKED-AFRWEAEHQKAFDELKVYLSSPHVMAPPIRGKPMKLYISATDGTIGSM 1008
PL RL ++D FRW AE + AF +LK + V+ + + +G +
Sbjct: 1228 --PLTRLTRKDFPFRWGAEQEAAFTKLKRLFAEEPVLRMFDPEAVTTVETDCSGFALGGV 1401
Query: 1009 LAQEDEDSKERAIFYLSRVLNDAETRYTMIEKLCLCLYFSCVK 1051
L QED + + S+ L+ AE Y + +K L ++ +C++
Sbjct: 1402 LTQEDGTGAAHPVAFHSQRLSPAEYNYPIHDKELLAVW-ACLR 1527
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 135 bits (340), Expect = 1e-31
Identities = 66/136 (48%), Positives = 97/136 (70%)
Frame = +2
Query: 797 EDVSKTAFRCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQVYIDDIVVKS 856
+D+ KTAF GTY + VMPFGLKNAG+TYQR++N +F D + M+VYIDD++VKS
Sbjct: 11 DDLEKTAFITDR--GTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVKS 184
Query: 857 PSRDDHLAHLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEINKNKAKAILD 916
DHL HL++ F+ + +Y +K+NP KC FGV +G+FLG++V ++GIE+N + AILD
Sbjct: 185 LRATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQGIEVNPKQITAILD 364
Query: 917 TSPPTSKKQLQSLLGK 932
P + +++Q L G+
Sbjct: 365 LPSPKNSREVQRLTGE 412
Score = 79.0 bits (193), Expect = 2e-14
Identities = 49/145 (33%), Positives = 79/145 (53%), Gaps = 7/145 (4%)
Frame = +3
Query: 914 ILDTSPPTSKKQLQSLL-------GKINFLRRFIANLSEKTKSFSPLLRLKKEDAFRWEA 966
IL SPP +Q + G+I L RFI+ ++K F LL K F W+
Sbjct: 336 ILSRSPPY*TSLVQRIAERSSDSRGRIAALNRFISRSTDKCLPFYKLLCGNKR--FVWDE 509
Query: 967 EHQKAFDELKVYLSSPHVMAPPIRGKPMKLYISATDGTIGSMLAQEDEDSKERAIFYLSR 1026
+ ++AF++LK YL++P V++ P G + LYI+ + + S+L +ED +++ IFY S+
Sbjct: 510 KCEEAFEQLKQYLTTPPVLSKPEAGDTLSLYIAISSTAVSSVLIREDR-GEQKPIFYTSK 686
Query: 1027 VLNDAETRYTMIEKLCLCLYFSCVK 1051
+ D ETRY +EK+ + S K
Sbjct: 687 RMTDPETRYPTLEKMAFAVITSARK 761
>BG586308 weakly similar to PIR|F84528|F8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 686
Score = 62.0 bits (149), Expect(2) = 9e-22
Identities = 29/70 (41%), Positives = 40/70 (56%)
Frame = -2
Query: 1473 GLPESLTTDQGTVFVGQKVASFAESWGIKLLNSTPYYAQANGQVEAANKTLISLIKKHVG 1532
GLP + TD G+ F+ K F E W I+L ++P Y Q+NGQ EA+NK +I +KK +
Sbjct: 682 GLPYEIVTDNGSHFISNKFREFCERWRIRLNTASPRYPQSNGQAEASNKIIIDGLKKRLD 503
Query: 1533 RKPKRWHQTL 1542
K W L
Sbjct: 502 LKKGCWADEL 473
Score = 61.6 bits (148), Expect(2) = 9e-22
Identities = 40/124 (32%), Positives = 67/124 (53%), Gaps = 3/124 (2%)
Frame = -3
Query: 1545 VLWAYRNSPKEATGATPFRLAYGQEAVLPAEVYLQS---CRIQRQEEIPSEDYWNMMLDE 1601
VLW++R +P+ AT +TPF +A+ EA+ PAEV + S R+ + E+ ++ +N
Sbjct: 465 VLWSHRTNPRGATKSTPFSMAHRVEAMAPAEVNVTSL*RSRMPQNIELNNDRLFN----A 298
Query: 1602 LVNLDEERLSALDILTRQKDRVAKAYNKKVRAKSFMPGDYVWKVVLPVDKRDKRYGKWAP 1661
L ++E R AL + + ++ YNK V+++ GD V V + ++ GK
Sbjct: 297 LETIEERRDQALLRIQNYQHQIESYYNKTVKSQPLKLGDIVLCKVFE-NTKELNAGKLGT 121
Query: 1662 NWEG 1665
NWEG
Sbjct: 120 NWEG 109
>CB065285 weakly similar to PIR|A84500|A84 probable retroelement gag/pol
polyprotein [imported] - Arabidopsis thaliana, partial
(2%)
Length = 592
Score = 96.7 bits (239), Expect = 7e-20
Identities = 66/171 (38%), Positives = 89/171 (51%), Gaps = 2/171 (1%)
Frame = -2
Query: 1130 WKLFFDGSSHKNGTGIGMFIVSPGGIPTKFKFRIKKNCSNNEAEYEALISGLEILIAFGA 1189
W L FDG+ + G GIG IVSP G F RI C+NN AEYEA I G+E I
Sbjct: 534 WGLVFDGAVNAYGKGIGAVIVSPQGHYIPFTARILFECTNNMAEYEACIFGIEEAIDMRI 355
Query: 1190 KNVVIKGDSELVIKQLTKEYKCVSENLARYYTKANNLLAKFDEARLSHVSRVDNQEANEL 1249
K++ I GDS LVI Q+ E++ NL Y A LL F + L H+ R +NQ A+ L
Sbjct: 354 KHLDIYGDSALVINQIKGEWETHHANLIPYRDYARRLLTYFTKVELHHIPRDENQMADAL 175
Query: 1250 AQIASGYMVDKCRLKELIEVKEKLNLSDLNIL--VIDNMAPNDWRKPIVDY 1298
A ++S + V+ +I+V+ S + + VID N +VDY
Sbjct: 174 ATLSSMFRVNHWNDVPIIKVQRLERPSHVFAIGNVIDQTGEN-----VVDY 37
>AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis thaliana
chromosome II BAC F26H6; putative retroelement pol
polyprotein, partial (1%)
Length = 636
Score = 91.3 bits (225), Expect = 3e-18
Identities = 53/167 (31%), Positives = 82/167 (48%), Gaps = 25/167 (14%)
Frame = -2
Query: 1040 KLCLCLYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSKPILHSRIGKWALALTEYSLTYAP 1099
K C L ++ +L++Y+ + S D IK++ KP L RI +W + L+EY + Y
Sbjct: 635 KTCCALAWAAKRLRHYMINHTTWLVSKMDPIKYIFEKPALTGRIARWQMLLSEYDIEYRS 456
Query: 1100 LKAVKGQAIADFLVDHTL----------PKEIVTYVGIQP---------------WKLFF 1134
KA+KG +AD L L P E + Y+ ++ W L F
Sbjct: 455 QKAIKGSILADHLAHQPLEDYRPIKFDFPDEEIMYLKMKDCDEPLFGEGPDPDSVWGLIF 276
Query: 1135 DGSSHKNGTGIGMFIVSPGGIPTKFKFRIKKNCSNNEAEYEALISGL 1181
DG+ + G GIG +++P G F R++ +C+NN AEYEA I G+
Sbjct: 275 DGAVNVYGNGIGAVLLTPKGAHIPFTARLRFDCTNNIAEYEACIMGI 135
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 90.1 bits (222), Expect = 7e-18
Identities = 56/179 (31%), Positives = 102/179 (56%)
Frame = +2
Query: 700 RFHPDVLVKIKEEIERLLKCKFIRTARYVDWLANVVPVIKKNGKMRVCIDFRDLNAATPK 759
R +P L +K +++ LL+ FI+ + Y + V+ + KK+G +R+ ID+ LN K
Sbjct: 92 RINPLKLKVLKLQLKDLLEKGFIQPSIYP*GVV-VLFLKKKDGFLRMSIDYPQLNNVNIK 268
Query: 760 DEYHMPVAEMMVDSTAGHEYLSLLDGYSGYNQIFIAEEDVSKTAFRCPGALGTYEWVVMP 819
+Y +P+ + + D+ G ++ +D G +Q + EDV KTAFR G YE +VM
Sbjct: 269 IKYPLPLIDELFDNLQGSKWFFKIDLRLG*HQHRVIGEDVPKTAFRI--RYGHYEILVMS 442
Query: 820 FGLKNAGATYQRVMNTIFHDFIETFMQVYIDDIVVKSPSRDDHLAHLRKSFERMRKYGL 878
FG N + +MN +F D++++ + V+ +DI++ S + ++H HLR + + ++ GL
Sbjct: 443 FG*TNPPMAFMELMNRVFQDYLDSLVIVFSNDILIYSKNENEHENHLRLALKVLKDIGL 619
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 86.7 bits (213), Expect = 7e-17
Identities = 52/188 (27%), Positives = 89/188 (46%)
Frame = +2
Query: 1324 LFKKNVDGTLLKCLSEDDAFIAISAVHDGLCGAHQAGIKMKWILFRQGMYWPTIMKDCME 1383
L+K+ D ++C++E++ + H H A K + + G +WPT+ KD
Sbjct: 41 LYKQCADNIYIRCVAEEEIPGILFHCHGSNYAGHFAVSKTVSKIQQAGFWWPTMFKDAHS 220
Query: 1384 YDKGCQDFQRHAGIQHVPASELHSIIKPWPFRGWALDLIGEINPCSSRQHKYIIVAIDYF 1443
+ C QR I + I++ F W +D +G SS +KYI+VA+DY
Sbjct: 221 FISKCDPCQRQGNIS*RNEMPQNFILEVEVFDVWGIDFMGPFP--SSYNNKYILVAVDYV 394
Query: 1444 TKWVEAIPLQNVTQDTVIDFIQNHIVYRFGLPESLTTDQGTVFVGQKVASFAESWGIKLL 1503
+KWVEAI V+ ++ I RFG+P + +D G+ F+ + + G++
Sbjct: 395 SKWVEAIASPTNDATVVVKMFKSVIFPRFGVPRVVISDGGSHFINKVFEKLLKKNGVRHK 574
Query: 1504 NSTPYYAQ 1511
+T Y+ Q
Sbjct: 575 VATAYHPQ 598
>AL367832 weakly similar to GP|14091845|gb Putative retroelement {Oryza
sativa}, partial (2%)
Length = 384
Score = 86.3 bits (212), Expect = 9e-17
Identities = 38/91 (41%), Positives = 62/91 (67%)
Frame = -1
Query: 652 QNEMVKLLKEFKDCFAWDYDEMPGLSRDLVELKLPIKEDKKPVKQLPRRFHPDVLVKIKE 711
+ ++ +LL+E+ D FA Y++MPGL +VE ++P K + PV+ RR HPD+ +KIK
Sbjct: 273 KRKIFQLLREYLDIFACSYEDMPGLDPKIVEHRIPTKPECPPVR*KLRRTHPDMALKIKS 94
Query: 712 EIERLLKCKFIRTARYVDWLANVVPVIKKNG 742
E+++ + F+ T Y +W+AN+VPV KK+G
Sbjct: 93 EVQKQIDAGFLMTVEYPEWVANIVPVPKKDG 1
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial (14%)
Length = 1708
Score = 77.0 bits (188), Expect = 6e-14
Identities = 72/301 (23%), Positives = 123/301 (39%), Gaps = 7/301 (2%)
Frame = +2
Query: 1350 HDGLCGAHQAGIKMKWILFRQGMYWPTIMKDCMEYDKGCQDFQRHAGIQHVPASELHSII 1409
HD H I+ R+ +WP + + + C G H+ +
Sbjct: 194 HDSTAAGHPGRNGTLEIVSRK-FFWPGQSQTVRRFVRNCDV----CGGIHIWRQAKRGFL 358
Query: 1410 KPWPFRG-----WALDLIGEINPCSSRQHKYIIVAIDYFTKWVEAIPLQNVTQDTVID-F 1463
KP P ++D I + P R +Y+ V +D +K V + + + F
Sbjct: 359 KPLPVPNRLHSDLSMDFITSLPPTRGRGSQYLWVIVDRLSKSVTLEEMDTMEAEACAQRF 538
Query: 1464 IQNHIVYRF-GLPESLTTDQGTVFVGQKVASFAESWGIKLLNSTPYYAQANGQVEAANKT 1522
+ H YRF G+P+S+ +D+G+ +VG+ F G+ L ST Y+ Q +G E N+
Sbjct: 539 LSCH--YRFHGMPQSIVSDRGSNWVGRFWREFCRLTGVTQLLSTSYHPQTDGGTERWNQE 712
Query: 1523 LISLIKKHVGRKPKRWHQTLGQVLWAYRNSPKEATGATPFRLAYGQEAVLPAEVYLQSCR 1582
+ ++++ +V W L V A RN + GATPF + +G V P +
Sbjct: 713 IQAVLRAYVCWSQDNWGDLLPTVQLALRNRHNSSIGATPFFVEHGYH-VDPIPTVEDTGG 889
Query: 1583 IQRQEEIPSEDYWNMMLDELVNLDEERLSALDILTRQKDRVAKAYNKKVRAKSFMPGDYV 1642
+ + E ++ M D + E I+ Q+ A A ++ A + GD V
Sbjct: 890 VVSEGEAAAQLLVKRMKDVTGFIQAE------IVAAQQRSEASANKRRCPADRYQVGDKV 1051
Query: 1643 W 1643
W
Sbjct: 1052 W 1054
>BG587127 weakly similar to PIR|H84506|H84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(13%)
Length = 415
Score = 65.1 bits (157), Expect = 2e-10
Identities = 41/125 (32%), Positives = 63/125 (49%)
Frame = +3
Query: 1146 GMFIVSPGGIPTKFKFRIKKNCSNNEAEYEALISGLEILIAFGAKNVVIKGDSELVIKQL 1205
G+ + SP K FR++ + SNNE YEALI+G+ + +N+ DS+LV Q
Sbjct: 3 GIRLTSPTNEILKQSFRLEFHASNNETNYEALIAGVRLAHGLKIRNIHAYCDSQLVASQF 182
Query: 1206 TKEYKCVSENLARYYTKANNLLAKFDEARLSHVSRVDNQEANELAQIASGYMVDKCRLKE 1265
+ EY+ E + Y L K D L+ + R +N +A+ LA +AS LK
Sbjct: 183 SGEYEARDELMDTYLKLVQKLAQKLDYFALTRIPRSENVQADALAALASS---SDPELKR 353
Query: 1266 LIEVK 1270
+I V+
Sbjct: 354 VIPVR 368
>TC83629 weakly similar to GP|6692261|gb|AAF24611.1| putative RNase H
{Arabidopsis thaliana}, partial (21%)
Length = 1071
Score = 53.1 bits (126), Expect = 9e-07
Identities = 33/92 (35%), Positives = 45/92 (48%), Gaps = 4/92 (4%)
Frame = +2
Query: 1168 SNNEAEYEALISGLEILIAFGAKNVVIKGDSELVIKQLTKEYKCVSENLARYYTKANNLL 1227
+ AEY AL+ GL+ G K V KGDSELVI Q+ +K E+L + +A L
Sbjct: 674 TKESAEYRALLLGLKHASMKGFKYVTAKGDSELVINQILDPWKIKDEHLKKLCAEALELS 853
Query: 1228 AKFDEARLSHVSRVDN----QEANELAQIASG 1255
F R+ H+SR N + AN + G
Sbjct: 854 DNFHSFRIQHISRERNYGAVRNANRAINLTDG 949
>BG644720
Length = 678
Score = 45.1 bits (105), Expect(2) = 2e-06
Identities = 45/154 (29%), Positives = 69/154 (44%), Gaps = 11/154 (7%)
Frame = -3
Query: 1291 WRKPIVDY------LQNPVGTTDRKTKY--RAMSYVIMGNE-LFKKNVDGTLLKCLSEDD 1341
WR+PI+ Y L+NP R++ Y R ++ NE L+ +G LL CL E++
Sbjct: 463 WRQPIIIYMCYGIFLENP-----RRSTYIRRHTPHLH*YNETLYI*LFEGVLL*CLGEEE 299
Query: 1342 AFIAISAVHDGLCGAHQAGIKMKWILFRQGMYWPT--IMKDCMEYDKGCQDFQRHAGIQH 1399
A H +CG+H++ K+ + + R G YW T ++K + A
Sbjct: 298 TIQAFQ*AHSRVCGSHKS--KLYFHIKRMGYYWNT*IMLKGAL-----------LANFIQ 158
Query: 1400 VPASELHSIIKPWPFRGWALDLIGEINPCSSRQH 1433
P LH I F W L++ G + P SS H
Sbjct: 157 QPPEVLHPTITS*LFESWELNVFG-LFPKSSGGH 59
Score = 26.6 bits (57), Expect(2) = 2e-06
Identities = 10/18 (55%), Positives = 13/18 (71%)
Frame = -1
Query: 1435 YIIVAIDYFTKWVEAIPL 1452
YI+ A+ YF KWVE + L
Sbjct: 54 YILAAVYYF*KWVEVVAL 1
>CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product {Drosophila
melanogaster}, partial (20%)
Length = 192
Score = 47.4 bits (111), Expect = 5e-05
Identities = 22/57 (38%), Positives = 36/57 (62%)
Frame = +1
Query: 1000 ATDGTIGSMLAQEDEDSKERAIFYLSRVLNDAETRYTMIEKLCLCLYFSCVKLKYYI 1056
A D IG+ L+Q+DE++ E I Y SR+L AE YT++E+ CL ++ ++Y+
Sbjct: 1 APDWGIGAGLSQKDEENHEHPIAYASRLLTAAERNYTVVERECLAAIWAIRNFRHYL 171
>AW684517 similar to GP|9927273|dbj Similar to Arabidopsis thaliana
chromosome II BAC F26H6; putative retroelement pol
polyprotein, partial (1%)
Length = 488
Score = 43.9 bits (102), Expect = 5e-04
Identities = 20/37 (54%), Positives = 25/37 (67%)
Frame = +1
Query: 528 FIVVPSKANYNLLLGREWIHGVGAVPSTLHQRISIWK 564
F V+ A+Y+ LLGR WIH GAV STLHQ++ K
Sbjct: 7 FQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLKFVK 117
>AL366605
Length = 422
Score = 41.2 bits (95), Expect = 0.003
Identities = 22/72 (30%), Positives = 38/72 (52%), Gaps = 1/72 (1%)
Frame = -3
Query: 4 FPSSLTKNAFTWFTTLAPRSVHTWAQLERIFHEQF-FRGECKVSLKDLASVKRKPAESID 62
F SL ++A W+T+L+ +HT+ +L F + F K + + L S+ +K ES
Sbjct: 216 FQDSLMEDAAEWYTSLSKNDIHTFDELAAAFKSHYGFNTRLKPNREFLRSLSQKKEESFR 37
Query: 63 DYLNRFRMLKSR 74
+Y R+R +R
Sbjct: 36 EYAQRWRGAAAR 1
>TC91935
Length = 1144
Score = 40.8 bits (94), Expect = 0.005
Identities = 17/40 (42%), Positives = 25/40 (62%)
Frame = +3
Query: 1015 DSKERAIFYLSRVLNDAETRYTMIEKLCLCLYFSCVKLKY 1054
D + A +YLS +L D T+YT I+KLCL ++C + Y
Sbjct: 984 DGTKIAAYYLSWILKDVGTKYTHIDKLCLSFLYACNNISY 1103
>BG644741
Length = 735
Score = 40.0 bits (92), Expect = 0.008
Identities = 25/85 (29%), Positives = 40/85 (46%), Gaps = 1/85 (1%)
Frame = -2
Query: 1 MKYFPSSLTKNAFTWFTTLAPRSVHTWAQLERIFHEQFFRGECKVSLKD-LASVKRKPAE 59
++ FP SL A WFT L S+ TW QL +F +++ K++ D + + P E
Sbjct: 587 LRVFPLSLMGEADIWFTELPYNSIFTWNQLRDVFLARYYPVSKKLNHNDRVNNFVALPGE 408
Query: 60 SIDDYLNRFRMLKSRCFTHVPEHEL 84
S+ +RF + VP H +
Sbjct: 407 SVSSSWDRF----TSFLRSVPNHRI 345
>TC91086
Length = 697
Score = 38.5 bits (88), Expect = 0.023
Identities = 13/22 (59%), Positives = 21/22 (95%)
Frame = -1
Query: 721 FIRTARYVDWLANVVPVIKKNG 742
FI+T+RY+ WL+N++P+IKK+G
Sbjct: 676 FIQTSRYIKWLSNILPIIKKHG 611
>BG448304
Length = 637
Score = 36.6 bits (83), Expect = 0.086
Identities = 18/55 (32%), Positives = 28/55 (50%), Gaps = 3/55 (5%)
Frame = +2
Query: 162 KLPSPEQR--KGKKYCKYHNFFGHWTSQCVRFRDLVQGALDSGRLKRL-KYPHEE 213
K P+ E + KYC++H H T C+ +D ++ + GRL +L K P E
Sbjct: 68 KTPTQENKGTDKTKYCRFHKCHRHLTDDCIHLKDTIEILIQRGRLNQLTKNPEPE 232
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 36.2 bits (82), Expect = 0.11
Identities = 15/36 (41%), Positives = 23/36 (63%)
Frame = -1
Query: 737 VIKKNGKMRVCIDFRDLNAATPKDEYHMPVAEMMVD 772
V KK+G R+CID+R N T K++Y +P + + D
Sbjct: 223 VRKKDGYFRMCIDYRQFNKVTTKNKYPLPRIDNLFD 116
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.137 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 52,669,854
Number of Sequences: 36976
Number of extensions: 754635
Number of successful extensions: 3929
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 3764
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3895
length of query: 1710
length of database: 9,014,727
effective HSP length: 110
effective length of query: 1600
effective length of database: 4,947,367
effective search space: 7915787200
effective search space used: 7915787200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0026.5


