

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0026.11
(461 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
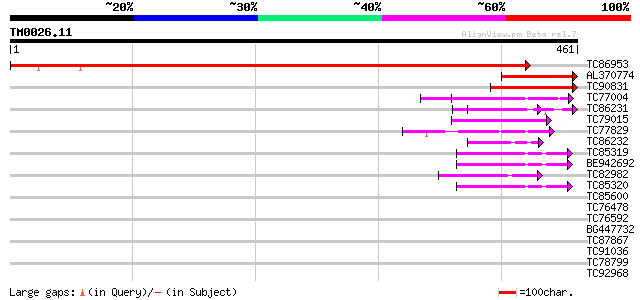
Score E
Sequences producing significant alignments: (bits) Value
TC86953 similar to GP|18252504|gb|AAL66290.1 adenosine 5'-phosph... 648 0.0
AL370774 similar to GP|18252504|gb| adenosine 5'-phosphosulfate ... 114 1e-25
TC90831 similar to GP|12831474|gb|AAB05871.2 PAPS-reductase-like... 98 6e-21
TC77004 homologue to SP|P38661|PDA6_MEDSA Probable protein disul... 53 2e-07
TC86231 similar to GP|21593313|gb|AAM65262.1 protein disulfide i... 47 1e-05
TC79015 weakly similar to PIR|F96562|F96562 hypothetical protein... 47 2e-05
TC77829 similar to GP|7211992|gb|AAF40463.1| Strong simialrity t... 45 5e-05
TC86232 similar to GP|21593313|gb|AAM65262.1 protein disulfide i... 43 3e-04
TC85319 similar to GP|8778824|gb|AAF79823.1| T6D22.5 {Arabidopsi... 42 7e-04
BE942692 similar to GP|8778824|gb| T6D22.5 {Arabidopsis thaliana... 42 7e-04
TC82982 weakly similar to GP|1388082|gb|AAC49355.1| thioredoxin ... 42 7e-04
TC85320 similar to GP|1532175|gb|AAB07885.1| similar to protein ... 42 7e-04
TC85600 homologue to SP|P29828|PDI_MEDSA Protein disulfide isome... 41 0.001
TC76478 similar to GP|14423558|gb|AAK62461.1 putative protein {A... 35 0.047
TC76592 similar to GP|1388088|gb|AAC49358.1| thioredoxin m {Pisu... 35 0.047
BG447732 homologue to GP|9837385|gb|A retinitis pigmentosa GTPas... 35 0.061
TC87867 similar to GP|15594012|emb|CAC69854. putative thioredoxi... 35 0.080
TC91036 weakly similar to GP|14423498|gb|AAK62431.1 Unknown prot... 34 0.14
TC78799 weakly similar to SP|P96132|THIO_THIRO Thioredoxin (TRX)... 32 0.67
TC92968 GP|20196877|gb|AAM14816.1 Expressed protein {Arabidopsis... 31 1.2
>TC86953 similar to GP|18252504|gb|AAL66290.1 adenosine 5'-phosphosulfate
reductase {Glycine max}, partial (89%)
Length = 1334
Score = 648 bits (1671), Expect = 0.0
Identities = 327/431 (75%), Positives = 366/431 (84%), Gaps = 8/431 (1%)
Frame = +3
Query: 1 MALAVTCSISISSSSSSSSTFQ---SSQPKFSQIASIRVSEIPHGGGV-NASQRRSFVKP 56
MALAVT S S++++ S+S + SS+ K QI S+RV E GV N QRR VK
Sbjct: 42 MALAVTSSSSVAATKSNSFFSRIGSSSESKVLQIGSLRVLERNVSSGVFNLPQRRYLVKA 221
Query: 57 ---PQRSRDSIAPLAATIVASDVAETEQDNYQQLAVDLENASPLQIMDAALEKFGNHIAI 113
+ DSI PLAATIVA ++ E E+++++QLA +LENASPL+IMD ALEKFGN IAI
Sbjct: 222 LNAESQRNDSIVPLAATIVAPEIIEKEEEDFEQLAKNLENASPLEIMDKALEKFGNDIAI 401
Query: 114 AFSGAEDVALIEYARLTGRPFRVFSLDTGRLNPETYRFFDEVEKHYGIHIEYMFPDAVEV 173
AFSGAEDVALIEYA LTGRP+RVFSLDTGRLNPETYR FD VEK YGI IEYMFPDAVEV
Sbjct: 402 AFSGAEDVALIEYAHLTGRPYRVFSLDTGRLNPETYRLFDAVEKRYGIRIEYMFPDAVEV 581
Query: 174 QGLVRSKGLFSFYEDGHQECCRVRKVRPLRRALKGLRAWITGQRKDQSPGTRSEVPVVQV 233
QGLVR+KGLFSFYEDGHQECCRVRKVRPLRRALKGLRAWITGQRKDQSPGTRSEVPVVQV
Sbjct: 582 QGLVRTKGLFSFYEDGHQECCRVRKVRPLRRALKGLRAWITGQRKDQSPGTRSEVPVVQV 761
Query: 234 DPVFEGVDGGVGSLVKWNPVANVKGHDIWSFLRTMNVPVNSLHSKGYISIGCEPCTRAVL 293
DPVFEG+DGG GSLVKWNPVANVKG+DIW+FLRTM+VPVN+LH++GY+SIGCEPCTR VL
Sbjct: 762 DPVFEGLDGGNGSLVKWNPVANVKGNDIWTFLRTMDVPVNALHAQGYVSIGCEPCTRPVL 941
Query: 294 PGQHEREGRWWWEDAKAKECGLHKGNVKQDAEAEL-NGNGVANTNGTATVADIFNTQNVV 352
PGQHEREGRWWWEDAKAKECGLHKGN+KQD A+ NGNGVA+ NG+A VADIF TQNVV
Sbjct: 942 PGQHEREGRWWWEDAKAKECGLHKGNLKQDDAAQNGNGNGVAHANGSAAVADIFTTQNVV 1121
Query: 353 SLSRTGIENLTKLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADG 412
SLSR GIENL KLE+ KEPW+VVLYAPWC +CQAMEESYV+LA L SGVKVGKF ADG
Sbjct: 1122SLSRAGIENLAKLEDPKEPWIVVLYAPWCRFCQAMEESYVNLAXNLXGSGVKVGKFXADG 1301
Query: 413 EQKEFAKRELG 423
++K + K ELG
Sbjct: 1302DEKXYPKSELG 1334
>AL370774 similar to GP|18252504|gb| adenosine 5'-phosphosulfate reductase
{Glycine max}, partial (12%)
Length = 483
Score = 114 bits (284), Expect = 1e-25
Identities = 53/61 (86%), Positives = 59/61 (95%)
Frame = +1
Query: 401 SGVKVGKFRADGEQKEFAKRELGLGSFPTIMFFPKHSSRPIKYPSENRDVDSLMAFVNAL 460
SGVKVGKFRADG++KE+AK ELGLGSFPTI+FFPKHSSRPIKYPSE RDVDSL+AFVNAL
Sbjct: 4 SGVKVGKFRADGDEKEYAKSELGLGSFPTILFFPKHSSRPIKYPSEKRDVDSLLAFVNAL 183
Query: 461 R 461
+
Sbjct: 184 K 186
>TC90831 similar to GP|12831474|gb|AAB05871.2 PAPS-reductase-like protein
{Catharanthus roseus}, partial (14%)
Length = 856
Score = 98.2 bits (243), Expect = 6e-21
Identities = 51/70 (72%), Positives = 55/70 (77%)
Frame = +1
Query: 392 VDLADKLAESGVKVGKFRADGEQKEFAKRELGLGSFPTIMFFPKHSSRPIKYPSENRDVD 451
VDLA+ LA SGVKVGKFRADG+ KE AK ELG + P FPKH S PIKYPSE R+VD
Sbjct: 331 VDLAENLAGSGVKVGKFRADGDGKECAKSELGWEATPQYSSFPKHCSWPIKYPSEKREVD 510
Query: 452 SLMAFVNALR 461
SLMAFVNALR
Sbjct: 511 SLMAFVNALR 540
>TC77004 homologue to SP|P38661|PDA6_MEDSA Probable protein disulfide
isomerase A6 precursor (EC 5.3.4.1) (P5). [Alfalfa]
{Medicago sativa}, complete
Length = 1575
Score = 53.1 bits (126), Expect = 2e-07
Identities = 33/100 (33%), Positives = 49/100 (49%), Gaps = 1/100 (1%)
Frame = +2
Query: 360 ENLTKLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAES-GVKVGKFRADGEQKEFA 418
EN K + + LV YAPWC +C+ + Y L + ++ V + K D + +
Sbjct: 200 ENFEKEVGQDKGALVEFYAPWCGHCKKLAPEYEKLGNSFKKAKSVLIAKVDCDEHKGVCS 379
Query: 419 KRELGLGSFPTIMFFPKHSSRPIKYPSENRDVDSLMAFVN 458
K G+ +PTI +FPK S P K+ R +SL FVN
Sbjct: 380 K--YGVSGYPTIQWFPKGSLEPKKFEGP-RTAESLAEFVN 490
Score = 50.4 bits (119), Expect = 1e-06
Identities = 38/125 (30%), Positives = 61/125 (48%), Gaps = 1/125 (0%)
Frame = +2
Query: 335 NTNGTATVADIFNTQNVVSLSRTGIENLTKLENRKEPWLVVLYAPWCPYCQAMEESYVDL 394
NT G V +VV L+ + L+ K+ LV YAPWC +C+++ Y +
Sbjct: 488 NTEGGTNVKIATAPSHVVVLTPETFNEVV-LDETKDV-LVEFYAPWCGHCKSLAPIYEKV 661
Query: 395 ADKL-AESGVKVGKFRADGEQKEFAKRELGLGSFPTIMFFPKHSSRPIKYPSENRDVDSL 453
A +E V + AD + ++ A++ + FPT+ FFPK + Y RD+D
Sbjct: 662 AAVFKSEDDVVIANLDAD-KYRDLAEK-YDVSGFPTLKFFPKGNKAGEDY-GGGRDLDDF 832
Query: 454 MAFVN 458
+AF+N
Sbjct: 833 VAFIN 847
>TC86231 similar to GP|21593313|gb|AAM65262.1 protein disulfide isomerase
precursor-like {Arabidopsis thaliana}, partial (79%)
Length = 2185
Score = 47.4 bits (111), Expect = 1e-05
Identities = 25/73 (34%), Positives = 36/73 (49%)
Frame = +1
Query: 361 NLTKLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQKEFAKR 420
N T + + +V YAPWC +CQA+ Y A +L + GV + K A E +
Sbjct: 385 NFTTVIENNQFVMVEFYAPWCGHCQALAPEYAAAATELKKDGVVLAKVDASVENE--LAY 558
Query: 421 ELGLGSFPTIMFF 433
E + FPT+ FF
Sbjct: 559 EYNVQGFPTVYFF 597
Score = 45.8 bits (107), Expect = 3e-05
Identities = 33/93 (35%), Positives = 50/93 (53%), Gaps = 4/93 (4%)
Frame = +1
Query: 373 LVVLYAPWCPYCQAMEESYVDLADKL-AESGVKVGKFRADGEQKEFAKRELGLGSFPTIM 431
L+ +YAPWC +CQA+E ++ LA L + + V K + AK + FPT++
Sbjct: 1438 LLEVYAPWCGHCQALEPTFNKLAKHLHSIESIVVAKMDGTTNEHPRAKSD----GFPTLL 1605
Query: 432 FFP--KHSSRPIKYPSENRDVD-SLMAFVNALR 461
F+P K SS PI DVD +++AF L+
Sbjct: 1606 FYPAGKKSSDPITV-----DVDRTVVAFYKFLK 1689
>TC79015 weakly similar to PIR|F96562|F96562 hypothetical protein F19K6.17
[imported] - Arabidopsis thaliana, partial (33%)
Length = 1200
Score = 47.0 bits (110), Expect = 2e-05
Identities = 25/81 (30%), Positives = 42/81 (50%)
Frame = +3
Query: 360 ENLTKLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQKEFAK 419
EN ++ N E LV+ YAPWC + + + A+ L E G + + DG++ A
Sbjct: 393 ENTERIVNGYEFVLVLGYAPWCSRSAELMPHFAEAANSLKEFGNSLVLAKLDGDRFTKAA 572
Query: 420 RELGLGSFPTIMFFPKHSSRP 440
LG+ +PT++ F +S+P
Sbjct: 573 SFLGIKGYPTLLLFVNGTSQP 635
>TC77829 similar to GP|7211992|gb|AAF40463.1| Strong simialrity to the
disulfide isomerase precursor homolog T21L14.14
GP|2702281 from A., partial (89%)
Length = 1646
Score = 45.4 bits (106), Expect = 5e-05
Identities = 33/128 (25%), Positives = 55/128 (42%), Gaps = 4/128 (3%)
Frame = +2
Query: 320 VKQDAEAELNGNGVANTN----GTATVADIFNTQNVVSLSRTGIENLTKLENRKEPWLVV 375
VK + LNG +N TA+ + N+ N L + KE W+V
Sbjct: 425 VKALLKERLNGKATGGSNEKKESTASSSVELNSSNFDEL----------VIKSKELWIVE 574
Query: 376 LYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQKEFAKRELGLGSFPTIMFFPK 435
+APWC +C+ + + ++ L + VK+G D ++ ++ + FPTI+ F
Sbjct: 575 FFAPWCGHCKKLAPEWKRASNNL-KGKVKLGHVDCDADKSLMSR--FNVQGFPTILVFGA 745
Query: 436 HSSRPIKY 443
PI Y
Sbjct: 746 DKDTPIPY 769
Score = 38.9 bits (89), Expect = 0.004
Identities = 29/94 (30%), Positives = 43/94 (44%)
Frame = +2
Query: 363 TKLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQKEFAKREL 422
+K+ N LV +APWC +C+A+ + + A + + V V AD Q +E
Sbjct: 146 SKVLNSNGVVLVEFFAPWCGHCKALTPIW-EKAATVLKGVVTVAALDADAHQS--LAQEY 316
Query: 423 GLGSFPTIMFFPKHSSRPIKYPSENRDVDSLMAF 456
G+ FPTI F P+ Y RDV + F
Sbjct: 317 GIRGFPTIKVF-SPGKPPVDYQGA-RDVKPIAEF 412
>TC86232 similar to GP|21593313|gb|AAM65262.1 protein disulfide isomerase
precursor-like {Arabidopsis thaliana}, partial (50%)
Length = 1315
Score = 42.7 bits (99), Expect = 3e-04
Identities = 24/63 (38%), Positives = 34/63 (53%), Gaps = 1/63 (1%)
Frame = +3
Query: 373 LVVLYAPWCPYCQAMEESYVDLADKLAE-SGVKVGKFRADGEQKEFAKRELGLGSFPTIM 431
L+ +YAPWC +CQ +E Y LA L + + K DG Q E + + FPT++
Sbjct: 612 LLEIYAPWCGHCQHLEPIYNKLAKHLRSIDSLVIAKM--DGTQNEHPRAK--SDGFPTLL 779
Query: 432 FFP 434
FFP
Sbjct: 780 FFP 788
>TC85319 similar to GP|8778824|gb|AAF79823.1| T6D22.5 {Arabidopsis
thaliana}, partial (62%)
Length = 881
Score = 41.6 bits (96), Expect = 7e-04
Identities = 23/95 (24%), Positives = 46/95 (48%), Gaps = 1/95 (1%)
Frame = +1
Query: 364 KLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLA-ESGVKVGKFRADGEQKEFAKREL 422
K++ + W V PWC YC+ + + D+ + E+ +++G+ ++ +K +
Sbjct: 232 KIKEKDTAWFVKFCVPWCKYCKNLGSLWDDVGKAMENENEIEIGEVDCGTDKAVCSK--V 405
Query: 423 GLGSFPTIMFFPKHSSRPIKYPSENRDVDSLMAFV 457
+ S+PT F + + RD++SL AFV
Sbjct: 406 DIHSYPTFKVF--YDGEEVAKYQGKRDIESLKAFV 504
>BE942692 similar to GP|8778824|gb| T6D22.5 {Arabidopsis thaliana}, partial
(62%)
Length = 588
Score = 41.6 bits (96), Expect = 7e-04
Identities = 23/95 (24%), Positives = 46/95 (48%), Gaps = 1/95 (1%)
Frame = +1
Query: 364 KLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLA-ESGVKVGKFRADGEQKEFAKREL 422
K++ + W V PWC YC+ + + D+ + E+ +++G+ ++ +K +
Sbjct: 142 KIKEKDTAWFVKFCVPWCKYCKNLGSLWDDVGKAMENENEIEIGEVDCGTDKAVCSK--V 315
Query: 423 GLGSFPTIMFFPKHSSRPIKYPSENRDVDSLMAFV 457
+ S+PT F + + RD++SL AFV
Sbjct: 316 DIHSYPTFKVF--YDGEEVAKYQGKRDIESLKAFV 414
>TC82982 weakly similar to GP|1388082|gb|AAC49355.1| thioredoxin h
{Arabidopsis thaliana}, partial (38%)
Length = 481
Score = 41.6 bits (96), Expect = 7e-04
Identities = 19/85 (22%), Positives = 44/85 (51%)
Frame = +1
Query: 349 QNVVSLSRTGIENLTKLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKF 408
+N + + + ++ K N+ + +V A WCP C+ + + +LA++ + + +
Sbjct: 25 ENFIVIQKDNKDDFNKELNKGKLVIVDFTASWCPPCKMIAPIFKNLAEEYKKVAIFL--- 195
Query: 409 RADGEQKEFAKRELGLGSFPTIMFF 433
+ DG++ +E G+ ++PT FF
Sbjct: 196 KVDGDENPEITKEYGISAYPTFFFF 270
>TC85320 similar to GP|1532175|gb|AAB07885.1| similar to protein disulfide
isomerase {Arabidopsis thaliana}, partial (78%)
Length = 746
Score = 41.6 bits (96), Expect = 7e-04
Identities = 23/95 (24%), Positives = 46/95 (48%), Gaps = 1/95 (1%)
Frame = +1
Query: 364 KLENRKEPWLVVLYAPWCPYCQAMEESYVDLADKLA-ESGVKVGKFRADGEQKEFAKREL 422
K++ + W V PWC YC+ + + D+ + E+ +++G+ ++ +K +
Sbjct: 181 KIKEKDTAWFVKFCVPWCKYCKNLGSLWDDVGKAMENENEIEIGEVDCGTDKAVCSK--V 354
Query: 423 GLGSFPTIMFFPKHSSRPIKYPSENRDVDSLMAFV 457
+ S+PT F + + RD++SL AFV
Sbjct: 355 DIHSYPTFKVF--YDGEEVAKYQGKRDIESLKAFV 453
>TC85600 homologue to SP|P29828|PDI_MEDSA Protein disulfide isomerase
precursor (PDI) (EC 5.3.4.1). [Alfalfa] {Medicago
sativa}, complete
Length = 1915
Score = 40.8 bits (94), Expect = 0.001
Identities = 30/110 (27%), Positives = 52/110 (47%), Gaps = 3/110 (2%)
Frame = +2
Query: 351 VVSLSRTGIENLTKLENRKEPWLVV-LYAPWCPYCQAMEESYVDLADKLA--ESGVKVGK 407
V++L T + K K ++VV YAPWC +C+ + Y A L+ E V + K
Sbjct: 152 VLTLDNTNFHDTVK----KHDFIVVEFYAPWCGHCKKLAPEYEKAASILSTHEPPVVLAK 319
Query: 408 FRADGEQKEFAKRELGLGSFPTIMFFPKHSSRPIKYPSENRDVDSLMAFV 457
A+ E + E + FPTI F ++ + I+ R+ D ++ ++
Sbjct: 320 VDANEEHNKDLASENDVKGFPTIKIF-RNGGKNIQEYKGPREADGIVEYL 466
Score = 27.7 bits (60), Expect = 9.7
Identities = 17/85 (20%), Positives = 36/85 (42%)
Frame = +2
Query: 373 LVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQKEFAKRELGLGSFPTIMF 432
L+ YAPWC +C+ + ++A +S V + D + + +PT+ F
Sbjct: 1244 LIEFYAPWCGHCKQLAPILDEVAVSF-QSDADVVIAKLDATANDIPTDTFEVQGYPTLYF 1420
Query: 433 FPKHSSRPIKYPSENRDVDSLMAFV 457
+ +S + R + ++ F+
Sbjct: 1421 --RSASGKLSQYDGGRTKEDIIEFI 1489
>TC76478 similar to GP|14423558|gb|AAK62461.1 putative protein {Arabidopsis
thaliana}, partial (50%)
Length = 1061
Score = 35.4 bits (80), Expect = 0.047
Identities = 20/62 (32%), Positives = 28/62 (44%)
Frame = +2
Query: 223 GTRSEVPVVQVDPVFEGVDGGVGSLVKWNPVANVKGHDIWSFLRTMNVPVNSLHSKGYIS 282
G R P F G ++ NP+ + DIW+FL T + SL+ +GY S
Sbjct: 371 GVRIGDPTAVGQEQFSPSSPGWPPFMRVNPILDWSYRDIWAFLLTCEMKYCSLYDQGYTS 550
Query: 283 IG 284
IG
Sbjct: 551 IG 556
>TC76592 similar to GP|1388088|gb|AAC49358.1| thioredoxin m {Pisum sativum},
partial (87%)
Length = 912
Score = 35.4 bits (80), Expect = 0.047
Identities = 18/63 (28%), Positives = 33/63 (51%)
Frame = +1
Query: 371 PWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQKEFAKRELGLGSFPTI 430
P LV +APWC C+ + +LA + A K+ ++ + ++ + G+ S PT+
Sbjct: 373 PVLVDFWAPWCGPCRMIAPIIDELAKEYAG---KISCYKLNTDENPNIATKYGIRSIPTV 543
Query: 431 MFF 433
+FF
Sbjct: 544 LFF 552
>BG447732 homologue to GP|9837385|gb|A retinitis pigmentosa GTPase
regulator-like protein {Takifugu rubripes}, partial (2%)
Length = 466
Score = 35.0 bits (79), Expect = 0.061
Identities = 18/35 (51%), Positives = 22/35 (62%)
Frame = +2
Query: 5 VTCSISISSSSSSSSTFQSSQPKFSQIASIRVSEI 39
+ CS S SSSSSSSS+F S PK Q S +S +
Sbjct: 68 LACSSSSSSSSSSSSSFSSKSPKPHQFRSFSISPL 172
>TC87867 similar to GP|15594012|emb|CAC69854. putative thioredoxin m2 {Pisum
sativum}, partial (79%)
Length = 816
Score = 34.7 bits (78), Expect = 0.080
Identities = 18/63 (28%), Positives = 32/63 (50%)
Frame = +3
Query: 371 PWLVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQKEFAKRELGLGSFPTI 430
P LV +APWC C+ + +LA + A K+ ++ + ++ G+ S PT+
Sbjct: 315 PVLVEFWAPWCGPCRMIHPIIDELAKEYAG---KLKCYKLNTDESPSVATRYGIRSIPTV 485
Query: 431 MFF 433
+FF
Sbjct: 486 IFF 494
>TC91036 weakly similar to GP|14423498|gb|AAK62431.1 Unknown protein
{Arabidopsis thaliana}, partial (42%)
Length = 744
Score = 33.9 bits (76), Expect = 0.14
Identities = 17/61 (27%), Positives = 29/61 (46%)
Frame = +2
Query: 373 LVVLYAPWCPYCQAMEESYVDLADKLAESGVKVGKFRADGEQKEFAKRELGLGSFPTIMF 432
LV YAPWC +C+ + A LA + + D ++ R+ + ++PTI+
Sbjct: 188 LVDFYAPWCGHCKRLSPELDAAAPVLAALKEPILIAKVDADKHTSLARKHDVDAYPTILL 367
Query: 433 F 433
F
Sbjct: 368 F 370
>TC78799 weakly similar to SP|P96132|THIO_THIRO Thioredoxin (TRX)
(Fragment). {Thiocapsa roseopersicina}, partial (37%)
Length = 734
Score = 31.6 bits (70), Expect = 0.67
Identities = 18/61 (29%), Positives = 31/61 (50%), Gaps = 1/61 (1%)
Frame = +2
Query: 373 LVVLYAPWC-PYCQAMEESYVDLADKLAESGVKVGKFRADGEQKEFAKRELGLGSFPTIM 431
LV YAPWC C + V+LA+ A KV ++ + ++ + G+ + PT++
Sbjct: 260 LVEFYAPWCGQECINIHSIMVELANDYAG---KVDFYKLNIDENPYITNRYGIQNLPTVV 430
Query: 432 F 432
F
Sbjct: 431 F 433
>TC92968 GP|20196877|gb|AAM14816.1 Expressed protein {Arabidopsis thaliana},
partial (8%)
Length = 636
Score = 30.8 bits (68), Expect = 1.2
Identities = 20/40 (50%), Positives = 26/40 (65%)
Frame = +2
Query: 12 SSSSSSSSTFQSSQPKFSQIASIRVSEIPHGGGVNASQRR 51
SSSSSSSS+ SS P FS ++S+ S+I G S+RR
Sbjct: 131 SSSSSSSSSLSSSYPSFS-LSSVDASKIT-SGTTRVSRRR 244
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.134 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,555,301
Number of Sequences: 36976
Number of extensions: 187901
Number of successful extensions: 2017
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 1554
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1783
length of query: 461
length of database: 9,014,727
effective HSP length: 99
effective length of query: 362
effective length of database: 5,354,103
effective search space: 1938185286
effective search space used: 1938185286
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0026.11


