

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
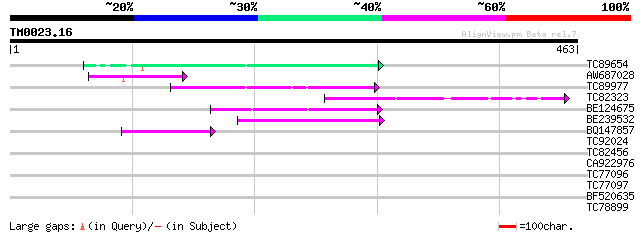
Query= TM0023.16
(463 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC89654 weakly similar to PIR|T05644|T05644 hypothetical protein... 80 1e-15
AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~... 77 1e-14
TC89977 similar to GP|5764395|gb|AAD51282.1| far-red impaired re... 55 4e-08
TC82323 similar to GP|7673677|gb|AAF66982.1| transposase {Zea ma... 54 1e-07
BE124675 weakly similar to GP|13365573|db hypothetical protein~s... 47 2e-05
BE239532 44 1e-04
BQ147857 weakly similar to GP|15983442|gb At1g10240/F14N23_12 {A... 41 9e-04
TC92024 similar to PIR|T05645|T05645 hypothetical protein F20D10... 39 0.004
TC82456 weakly similar to GP|18057159|gb|AAL58182.1 putative far... 38 0.007
CA922976 weakly similar to GP|9502168|gb|A contains simlarity to... 34 0.14
TC77096 similar to PIR|T05572|T05572 glucose-6-phosphate isomera... 31 0.89
TC77097 similar to PIR|T05572|T05572 glucose-6-phosphate isomera... 31 0.89
BF520635 weakly similar to GP|11994240|db gb|AAF40458.1~gene_id:... 28 5.7
TC78899 similar to GP|3900936|emb|CAA10167.1 glucan endo-1 3-bet... 28 7.5
>TC89654 weakly similar to PIR|T05644|T05644 hypothetical protein F20D10.290
- Arabidopsis thaliana, partial (65%)
Length = 1358
Score = 80.5 bits (197), Expect = 1e-15
Identities = 67/256 (26%), Positives = 101/256 (39%), Gaps = 11/256 (4%)
Frame = +1
Query: 61 TEKYLCAWHLLRNANQNLGNPLLTKGFKNCMFRKKWDELVLNLGLQD-----------NC 109
+EK +WH+ NQ L LLT +N R L+++L L N
Sbjct: 547 SEKIEASWHIC--INQIL---LLTTNSRNVFMRV*---LLMSLSLVGAHYWKDTTSWINE 702
Query: 110 WVKETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQLQNCVK 169
W + Y R WV +LRG FF E HV++ L ++Q + V
Sbjct: 703 WPQSMYNARQHWVPVYLRGSFFGEIPLNDGNECLNFFFDGHVNASTTLQLLVRQYEKAVS 882
Query: 170 HMRFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFRPVLQHAPLMKVLDCTD 229
RE++ D + + P ++T +E A +T +F+ F+ L D
Sbjct: 883 TWHERELKADFETSNSNPVLRTP-SPMEKQAASLYTRKMFMKFQEELVGTMANPATKIED 1059
Query: 230 AVTCLNYTVAKLSFLAKEWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAHLRIEELS 289
+ T Y VAK V+ +S SC E G+ C HI+ V + L
Sbjct: 1060SGTITTYRVAKFGENQTSHTVAFNSSEMKASCSCQMFEYSGIVCRHILAVFRAKNVLTLP 1239
Query: 290 ECFIFKRWSKGAKDGV 305
++ KRW+ AK GV
Sbjct: 1240SHYVLKRWTMNAKTGV 1287
Score = 33.5 bits (75), Expect = 0.18
Identities = 14/54 (25%), Positives = 29/54 (52%)
Frame = +2
Query: 10 MQFMVELASKDDNYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFDVTEK 63
+ ++ + +++ +F +D N+ ++FW D R +Y +GD + FD T K
Sbjct: 134 LDYLKRMRAENHAFFYAVQSDVDNAGGNIFWADETCRTNYSYFGDTVIFDTTYK 295
>AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~similar to
Arabidopsis thaliana F20D10.300, partial (11%)
Length = 662
Score = 77.0 bits (188), Expect = 1e-14
Identities = 36/89 (40%), Positives = 49/89 (54%), Gaps = 8/89 (8%)
Frame = +1
Query: 65 LCAWHLLRNANQNLGNPLLTKGFKNCM--------FRKKWDELVLNLGLQDNCWVKETYE 116
LCAWHL +NA +N+ +GF+ M F W EL+ L+ N WV +TY
Sbjct: 262 LCAWHLHKNAQENIKKTPFLEGFRKAMYSNFTPEQFEDFWSELIQKNELEGNAWVIKTYA 441
Query: 117 KRSIWVAAHLRGKFFAGCRTTSRCEGFQS 145
+S+W A+LR KFF RTTS+CE +
Sbjct: 442 NKSLWATAYLRDKFFGRIRTTSQCEAINA 528
>TC89977 similar to GP|5764395|gb|AAD51282.1| far-red impaired response
protein {Arabidopsis thaliana}, partial (35%)
Length = 1523
Score = 55.5 bits (132), Expect = 4e-08
Identities = 44/172 (25%), Positives = 75/172 (43%), Gaps = 1/172 (0%)
Frame = +2
Query: 132 AGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQLQNCVKHMRFREIEDDCHSIHGVPAMKT 191
AG T R E S K++H + L +F++Q + + + E D ++H PA+K+
Sbjct: 2 AGTSTAQRSESMNSFFDKYIHKKITLKEFVKQYRLILLNRYDEEEIADFDTLHKQPALKS 181
Query: 192 NLESLEMSATKHFTNNVFLLFR-PVLQHAPLMKVLDCTDAVTCLNYTVAKLSFLAKEWHV 250
E + +T+ +F F+ VL A ++ D T + Y V +E+ V
Sbjct: 182 P-SPWEKQMSTIYTHAIFKKFQVEVLGVAGCQSRIEVGDG-TAVRYIVQDYE-KDEEFLV 352
Query: 251 SVSSDNKDFKYSCMKMESRGLACEHIVDVLAHLRIEELSECFIFKRWSKGAK 302
+ + + C E +G C H + VL + +I KRW+K AK
Sbjct: 353 TWKELSSEVSCFCRLFEYKGFLCRHALSVLQRCGCSSVPSHYIMKRWTKDAK 508
>TC82323 similar to GP|7673677|gb|AAF66982.1| transposase {Zea mays},
partial (2%)
Length = 1165
Score = 54.3 bits (129), Expect = 1e-07
Identities = 49/201 (24%), Positives = 85/201 (41%), Gaps = 1/201 (0%)
Frame = +1
Query: 258 DFKYSCMKMESRGLACEHIVDVLAHLRIEELSECFIFKRWSKGAK-DGVYSGKNIENDFW 316
+F+ SC ESRG+ C H+ V+ ++ + +C + RW+K AK + + S N E D
Sbjct: 22 EFQCSCRLFESRGIPCCHVFFVMKEEDVDHIPKCLVMTRWTKNAKGEFLNSDSNGEIDA- 198
Query: 317 DFQKSSRCSALMDLYRALGYLNSNTAADFNDATQKASELIEQSKAKKSCQREGSLSLSQT 376
+ + +R A + S F D + L KK C E S
Sbjct: 199 NMIELARFGAYCSAFTTFCKEASKKNGVFRDIMDEILNL-----QKKYCDVEDSTRSKSF 363
Query: 377 NILNLKDPIRLRRKGRGTKNKSSLGLTVKHVINCLIFKTPGHNKLTGTYCSQQGHTMSDF 436
+ DP+ ++ KG K K + +V+H C K+ H T +CS + S+
Sbjct: 364 TDDQVGDPVTVKSKG-APKKKKNAAKSVRHCSRC---KSTSH---TARHCSDTHN--SEL 516
Query: 437 EGGEDEINLFTVDLVVDEVSK 457
+ E+ +++ D + + K
Sbjct: 517 QYNEELVSISMEDSLSQKEKK 579
>BE124675 weakly similar to GP|13365573|db hypothetical protein~similar to
Arabidopsis thaliana F20D10.300, partial (4%)
Length = 448
Score = 47.0 bits (110), Expect = 2e-05
Identities = 37/140 (26%), Positives = 64/140 (45%)
Frame = +2
Query: 165 QNCVKHMRFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFRPVLQHAPLMKV 224
+ V+ R++EIE +P + N+ L+ A+ +T F +F+ + + + V
Sbjct: 5 ERIVEEQRYKEIEASDEMKGCLPKLMGNVVVLK-HASVAYTPRAFEVFQQRYEKSLNVIV 181
Query: 225 LDCTDAVTCLNYTVAKLSFLAKEWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAHLR 284
Y V A+++ V+ SS + SCMK E G C H + VL +
Sbjct: 182 NQHKRDGYLFEYKVNTYGH-ARQYTVTFSSSDNTVVCSCMKFEHVGFLCSHALKVLDNRN 358
Query: 285 IEELSECFIFKRWSKGAKDG 304
I+ + +I KRW+K A+ G
Sbjct: 359 IKVVPSRYILKRWTKDARLG 418
>BE239532
Length = 645
Score = 43.9 bits (102), Expect = 1e-04
Identities = 28/121 (23%), Positives = 52/121 (42%), Gaps = 1/121 (0%)
Frame = +3
Query: 187 PAMKTNLESLEMSATKHFTNNVFLLF-RPVLQHAPLMKVLDCTDAVTCLNYTVAKLSFLA 245
P + N+ A+ +T+ F +F L+ ++ + + N+ V
Sbjct: 15 PGLVINISETLRHASTVYTHKGFRIFLNEYLEGTGGSTSIEISQSNNLSNHEVMLNHKPN 194
Query: 246 KEWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAHLRIEELSECFIFKRWSKGAKDGV 305
K++ V+ S N SC K +S G+ C H + + I + + + KRWSK A+ +
Sbjct: 195 KKYAVTFDSSNMKINCSCRKFDSMGILCSHALRIYNIKGILRIPDQYFLKRWSKNARSVI 374
Query: 306 Y 306
Y
Sbjct: 375 Y 377
>BQ147857 weakly similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidopsis
thaliana}, partial (26%)
Length = 560
Score = 41.2 bits (95), Expect = 9e-04
Identities = 22/77 (28%), Positives = 35/77 (44%)
Frame = +1
Query: 92 FRKKWDELVLNLGLQDNCWVKETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHV 151
F K W ++V GL N + Y R+ W LR FFAG + + E + + +
Sbjct: 202 FEKSWRQMVDKYGLHANKHIISLYSLRTFWALPFLRRYFFAGLTSXCQTESINVFIQRFL 381
Query: 152 HSRYNLTDFLQQLQNCV 168
++ FLQQ+ + V
Sbjct: 382 SAQSQPXRFLQQVADIV 432
>TC92024 similar to PIR|T05645|T05645 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (11%)
Length = 938
Score = 38.9 bits (89), Expect = 0.004
Identities = 26/107 (24%), Positives = 47/107 (43%), Gaps = 5/107 (4%)
Frame = +3
Query: 262 SCMKMESRGLACEHIVDVLAHLRIEELSECFIFKRWSKGAKDGVYSGKNIENDFWDFQKS 321
SC E GL C HI+ V + L +I KRW+K AK V ++ + + + +S
Sbjct: 96 SCQMFEFSGLLCRHILAVFRVTNVLTLPSHYILKRWTKNAKSNVSLQEHSSHAYTYYLES 275
Query: 322 SRCSALMDLYRALGYLNSNTAAD-----FNDATQKASELIEQSKAKK 363
+ A +++ ++ DA Q+A++ + Q K+
Sbjct: 276 HTVRYNTLRHEAFKFVDKGASSPETYDVAKDALQEAAKRVAQVMRKE 416
>TC82456 weakly similar to GP|18057159|gb|AAL58182.1 putative far-red
impaired response protein {Oryza sativa}, partial (7%)
Length = 1244
Score = 38.1 bits (87), Expect = 0.007
Identities = 14/50 (28%), Positives = 27/50 (54%)
Frame = +2
Query: 12 FMVELASKDDNYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFDVT 61
F + ++ ++C D ++Q+LFW D R +Y+ +G+V+ D T
Sbjct: 737 FFHRMQKQNSQFYCAMDMDDKRNIQNLFWADARCRAAYEYFGEVITLDTT 886
>CA922976 weakly similar to GP|9502168|gb|A contains simlarity to Arabidopsis
thaliana far-red impaired response protein
(GB:AAD51282.1), partial (11%)
Length = 827
Score = 33.9 bits (76), Expect = 0.14
Identities = 17/65 (26%), Positives = 28/65 (42%)
Frame = -3
Query: 235 NYTVAKLSFLAKEWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAHLRIEELSECFIF 294
+Y + L E HV + + SC + ES G+ C H + +L +L E +
Sbjct: 780 SYIIRHSKKLDGERHVLWLPEKEQILCSCKEFESSGILCRHALRILVTKNYFQLPEKYYL 601
Query: 295 KRWSK 299
RW +
Sbjct: 600 SRWHR 586
>TC77096 similar to PIR|T05572|T05572 glucose-6-phosphate isomerase (EC
5.3.1.9) - Arabidopsis thaliana, partial (89%)
Length = 2494
Score = 31.2 bits (69), Expect = 0.89
Identities = 22/81 (27%), Positives = 39/81 (47%), Gaps = 2/81 (2%)
Frame = -1
Query: 109 CWVKETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLT--DFLQQLQN 166
C V ++Y +R I + LR K ++ CRT++ + S+ R NLT D + + N
Sbjct: 868 CVVNKSYLERRIVRSKCLRNKLWSKCRTSNPNRKYLSKPSLRG*RRLNLTRDDIISKTTN 689
Query: 167 CVKHMRFREIEDDCHSIHGVP 187
+K + ++ S G+P
Sbjct: 688 SIKGLINLRLKKYIRSTIGIP 626
>TC77097 similar to PIR|T05572|T05572 glucose-6-phosphate isomerase (EC
5.3.1.9) - Arabidopsis thaliana, partial (40%)
Length = 1017
Score = 31.2 bits (69), Expect = 0.89
Identities = 22/81 (27%), Positives = 39/81 (47%), Gaps = 2/81 (2%)
Frame = -3
Query: 109 CWVKETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLT--DFLQQLQN 166
C V ++Y +R I + LR K ++ CRT++ + S+ R NLT D + + N
Sbjct: 382 CVVNKSYLERRIVRSKCLRNKLWSKCRTSNPNRKYLSKPSLRG*RRLNLTRDDIISKTTN 203
Query: 167 CVKHMRFREIEDDCHSIHGVP 187
+K + ++ S G+P
Sbjct: 202 SIKGLINLRLKKYIRSTIGIP 140
>BF520635 weakly similar to GP|11994240|db
gb|AAF40458.1~gene_id:MUJ8.11~similar to unknown protein
{Arabidopsis thaliana}, partial (20%)
Length = 621
Score = 28.5 bits (62), Expect = 5.7
Identities = 13/29 (44%), Positives = 19/29 (64%), Gaps = 3/29 (10%)
Frame = -1
Query: 143 FQSELGKHVH---SRYNLTDFLQQLQNCV 168
FQS +G H SR N+T+F +Q++ CV
Sbjct: 378 FQSNIGTSSHEHFSRVNITEFYRQVKRCV 292
>TC78899 similar to GP|3900936|emb|CAA10167.1 glucan endo-1
3-beta-d-glucosidase {Cicer arietinum}, partial (94%)
Length = 1296
Score = 28.1 bits (61), Expect = 7.5
Identities = 15/42 (35%), Positives = 24/42 (56%)
Frame = -3
Query: 355 LIEQSKAKKSCQREGSLSLSQTNILNLKDPIRLRRKGRGTKN 396
L+ Q A + REG +SL+ + I ++ +P+R R G KN
Sbjct: 349 LLTQFVASFAFLREGRVSLATSRINSMLEPLRAWRAGSSGKN 224
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.135 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,045,050
Number of Sequences: 36976
Number of extensions: 247786
Number of successful extensions: 1143
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 1136
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1140
length of query: 463
length of database: 9,014,727
effective HSP length: 99
effective length of query: 364
effective length of database: 5,354,103
effective search space: 1948893492
effective search space used: 1948893492
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0023.16


