

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
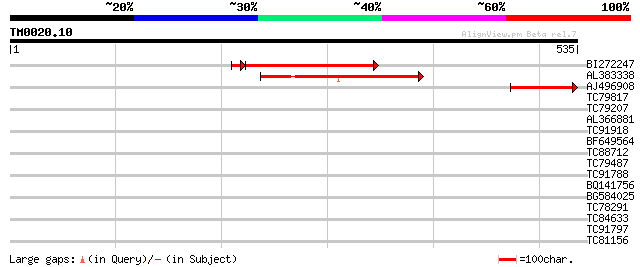
Query= TM0020.10
(535 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BI272247 similar to GP|7248400|dbj| ESTs AU082452(S2330) AU05813... 204 1e-54
AL383338 similar to GP|7248400|dbj| ESTs AU082452(S2330) AU05813... 179 2e-45
AJ496908 similar to GP|7293347|gb|A B-H1 gene product {Drosophil... 98 9e-21
TC79817 similar to GP|22795037|gb|AAN05420.1 putative RING prote... 40 0.002
TC79207 similar to GP|10086489|gb|AAG12549.1 Unknown Protein {Ar... 37 0.025
AL366881 similar to PIR|T00399|T003 probable AP2 domain transcri... 32 0.47
TC91918 homologue to GP|9369375|gb|AAF87124.1| F10A5.29 {Arabido... 30 1.8
BF649564 weakly similar to GP|21539507|gb unknown protein {Arabi... 30 2.3
TC88712 weakly similar to GP|9294466|dbj|BAB02685.1 gb|AAD14522.... 30 3.0
TC79487 similar to PIR|B86445|B86445 unknown protein [imported] ... 30 3.0
TC91788 auxin efflux carrier protein [Medicago truncatula] 30 3.0
BQ141756 similar to GP|19386784|dbj hypothetical protein {Oryza ... 29 4.0
BG584025 homologue to PIR|A46259|A46 recombination protein recA ... 29 5.2
TC78291 weakly similar to PIR|A84793|A84793 nodulin-like protein... 29 5.2
TC84633 similar to PIR|T49099|T49099 dihydrolipoamide S-acetyltr... 28 6.8
TC91797 similar to GP|5734493|emb|CAB52700.1 hypothetical protei... 28 6.8
TC81156 similar to PIR|T45754|T45754 hypothetical protein F24M12... 28 8.8
>BI272247 similar to GP|7248400|dbj| ESTs AU082452(S2330) AU058131(S5384)
D40386(S2330) D23841(R0349) correspond to a region of
the, partial (4%)
Length = 420
Score = 204 bits (518), Expect(2) = 1e-54
Identities = 98/126 (77%), Positives = 111/126 (87%)
Frame = +1
Query: 223 GPTLKRSQFPIRQGISSKLSLHATAAGCLGSALISAFTYHMIRELNVNEPDIMSLWIVSI 282
G TLK++QF IRQG+SS+LSLHATAAGC G ALIS+FTYHMI EL+ N+ DIMSLW+VSI
Sbjct: 43 GTTLKKAQFFIRQGVSSRLSLHATAAGCFGGALISSFTYHMIHELDENKSDIMSLWVVSI 222
Query: 283 FSGLIWLVGIFHVATAIGRTTDEISFSSRLHPFSVFKYPHAIGALASVFISSFITMTIFT 342
FSGLIWLVGI H+ATA RTTD ISFSSRLHPFS+FKYPHAIG LASVF++SF TM IF
Sbjct: 223 FSGLIWLVGILHLATATSRTTDSISFSSRLHPFSIFKYPHAIGGLASVFLASFTTMAIFM 402
Query: 343 GGVVFI 348
GGV+FI
Sbjct: 403 GGVIFI 420
Score = 28.1 bits (61), Expect(2) = 1e-54
Identities = 11/13 (84%), Positives = 12/13 (91%)
Frame = +3
Query: 210 HTHHLGLMIRSFT 222
HT HLGLMIR+FT
Sbjct: 3 HTQHLGLMIRAFT 41
>AL383338 similar to GP|7248400|dbj| ESTs AU082452(S2330) AU058131(S5384)
D40386(S2330) D23841(R0349) correspond to a region of
the, partial (4%)
Length = 475
Score = 179 bits (454), Expect = 2e-45
Identities = 93/157 (59%), Positives = 115/157 (73%), Gaps = 3/157 (1%)
Frame = +2
Query: 237 ISSKLSLHATAAGCLGSALISAFTYHMIRELNVNEPDIMSLWIVSIFSGLIWLVGIFHVA 296
+SS SL+AT+ GCLGSA+IS+F YHM+RE ++ + +LWIVSIFSGLIWLVG+ HV
Sbjct: 11 VSSFFSLYATSFGCLGSAIISSFIYHMLRE--PDDRNYFTLWIVSIFSGLIWLVGVLHVV 184
Query: 297 TAIGRTT-DEISF--SSRLHPFSVFKYPHAIGALASVFISSFITMTIFTGGVVFIVGQLC 353
TAI RTT +S S +P S+ +P AIG L VF+SSF TM IF GGV+FIVG LC
Sbjct: 185 TAINRTTVSSVSLINKSMFYPCSLLXHPKAIGGLFGVFLSSFTTMCIFIGGVLFIVGNLC 364
Query: 354 IKPVHLLFFWLTYFLFPVFSLSLLQPLLRVIKMNSVK 390
I+P HLL+FWL YFLFP+ SL LL PL ++K NS K
Sbjct: 365 IRPPHLLYFWLIYFLFPLVSLPLLHPLQLLLKANSAK 475
>AJ496908 similar to GP|7293347|gb|A B-H1 gene product {Drosophila
melanogaster}, partial (2%)
Length = 210
Score = 97.8 bits (242), Expect = 9e-21
Identities = 51/64 (79%), Positives = 55/64 (85%), Gaps = 1/64 (1%)
Frame = +1
Query: 473 SVSPGNIRTSFGAAFCTAIAGIVVLLFGNISDVGGAVAAGHVRDDSERSS-AVAGLDTKE 531
SV PG IRTSFG AFCTAIAGIVVLLFGNISD GGAVAAG+V DDSERSS V+GLD+KE
Sbjct: 1 SVGPGRIRTSFGVAFCTAIAGIVVLLFGNISDAGGAVAAGNVSDDSERSSPVVSGLDSKE 180
Query: 532 SVHV 535
+ V
Sbjct: 181 AARV 192
>TC79817 similar to GP|22795037|gb|AAN05420.1 putative RING protein {Populus
x canescens}, partial (32%)
Length = 1228
Score = 40.4 bits (93), Expect = 0.002
Identities = 33/116 (28%), Positives = 54/116 (46%), Gaps = 10/116 (8%)
Frame = +2
Query: 337 TMTIFTGGVVFIVGQLCIKPV------HLLFFWLTYFLFPVFSLSL--LQPLLRVIKMNS 388
++ FT G VF G LC P+ H + F +L P +SLS+ +P L VI +
Sbjct: 368 SVVTFTAGPVFTNGSLCKVPLLHLMSHHSVQFARMAYLTPKWSLSMAGARPSLAVIATVT 547
Query: 389 VKMQIVGFLFSL--ISSGFGFYYGHSHWKWGHLVVFSVIQSTGTGILHAFSRVLVL 442
+ + FL+ L + GF Y W+W H + V+ + IL + +++L
Sbjct: 548 QNLPLKIFLYRLDHLHQGFNLY-----WQWQHRLKMEVVSNFHIVILIRLNTLILL 700
>TC79207 similar to GP|10086489|gb|AAG12549.1 Unknown Protein {Arabidopsis
thaliana}, partial (20%)
Length = 1719
Score = 36.6 bits (83), Expect = 0.025
Identities = 17/44 (38%), Positives = 26/44 (58%)
Frame = -1
Query: 156 LSFHLDGHFPKFITVASTAVGVFFCLPAGFFKVTAIFIPYIAGI 199
LSFHL+ HFP+F++ + F P F+VT + +PYI +
Sbjct: 945 LSFHLEAHFPEFLSYFANG-SAQFQTPGHVFQVTYMTLPYILAV 817
>AL366881 similar to PIR|T00399|T003 probable AP2 domain transcription factor
[imported] - Arabidopsis thaliana, partial (26%)
Length = 477
Score = 32.3 bits (72), Expect = 0.47
Identities = 19/61 (31%), Positives = 29/61 (47%), Gaps = 1/61 (1%)
Frame = -1
Query: 351 QLCIKPVHLLFFWLTYFLFPVFSLS-LLQPLLRVIKMNSVKMQIVGFLFSLISSGFGFYY 409
++C KP L WLT F +P S + RV + + + +GF S++ FGF
Sbjct: 348 RVCSKPYS*LLSWLTNFRYPFSPCSHSYTSICRVFCVIFISLSFLGFFISILRIIFGF*I 169
Query: 410 G 410
G
Sbjct: 168 G 166
>TC91918 homologue to GP|9369375|gb|AAF87124.1| F10A5.29 {Arabidopsis
thaliana}, partial (38%)
Length = 1171
Score = 30.4 bits (67), Expect = 1.8
Identities = 14/42 (33%), Positives = 23/42 (54%)
Frame = -1
Query: 301 RTTDEISFSSRLHPFSVFKYPHAIGALASVFISSFITMTIFT 342
+T+ I F++ HPF+ PH + +F SSF + T F+
Sbjct: 199 KTSTTIRFANFSHPFTPCSLPHTFVSATCIFRSSFTSHTPFS 74
>BF649564 weakly similar to GP|21539507|gb unknown protein {Arabidopsis
thaliana}, partial (44%)
Length = 647
Score = 30.0 bits (66), Expect = 2.3
Identities = 13/32 (40%), Positives = 19/32 (58%)
Frame = +2
Query: 354 IKPVHLLFFWLTYFLFPVFSLSLLQPLLRVIK 385
I LL FW+ YFLF V + L+PL +++
Sbjct: 68 ISLAELLRFWVCYFLFLVLPILFLKPLFLILE 163
>TC88712 weakly similar to GP|9294466|dbj|BAB02685.1
gb|AAD14522.1~gene_id:MGF10.2~similar to unknown protein
{Arabidopsis thaliana}, partial (48%)
Length = 746
Score = 29.6 bits (65), Expect = 3.0
Identities = 12/27 (44%), Positives = 18/27 (66%)
Frame = +2
Query: 359 LLFFWLTYFLFPVFSLSLLQPLLRVIK 385
LL FW+ YFLF V + L+PL +++
Sbjct: 80 LLRFWVCYFLFLVLPILFLKPLFLILE 160
>TC79487 similar to PIR|B86445|B86445 unknown protein [imported] -
Arabidopsis thaliana, partial (46%)
Length = 921
Score = 29.6 bits (65), Expect = 3.0
Identities = 14/45 (31%), Positives = 24/45 (53%), Gaps = 1/45 (2%)
Frame = +2
Query: 463 AGLLVGFTVGSVSPGNIRTSFGAAFCTAIAGIV-VLLFGNISDVG 506
A ++G+ VGS P N++ F C A++ ++ FG S +G
Sbjct: 5 AATVLGYMVGSGLPSNVKKVFHPIICCALSAVLTAFAFGYFSKLG 139
>TC91788 auxin efflux carrier protein [Medicago truncatula]
Length = 2076
Score = 29.6 bits (65), Expect = 3.0
Identities = 19/65 (29%), Positives = 30/65 (45%), Gaps = 6/65 (9%)
Frame = -2
Query: 331 FISSFITMTIFTGGVVFIVGQLCI------KPVHLLFFWLTYFLFPVFSLSLLQPLLRVI 384
F+ SF+ + V+ + G +C+ P +FFWL Y F S+S L +
Sbjct: 1436 FLMSFLQTMMRISLVMTLAGGICVFLLTSPPPSKSIFFWLIYGDFTASSISTSLSLGLMF 1257
Query: 385 KMNSV 389
+NSV
Sbjct: 1256 SINSV 1242
>BQ141756 similar to GP|19386784|dbj hypothetical protein {Oryza sativa
(japonica cultivar-group)}, partial (7%)
Length = 1219
Score = 29.3 bits (64), Expect = 4.0
Identities = 15/34 (44%), Positives = 23/34 (67%)
Frame = -2
Query: 306 ISFSSRLHPFSVFKYPHAIGALASVFISSFITMT 339
+SFSS+ HPF F + + ALAS+ +S F+ +T
Sbjct: 636 LSFSSQYHPF-FFSFILFLTALASICLSYFLCLT 538
>BG584025 homologue to PIR|A46259|A46 recombination protein recA homolog -
Arabidopsis thaliana (fragment), partial (43%)
Length = 828
Score = 28.9 bits (63), Expect = 5.2
Identities = 18/60 (30%), Positives = 30/60 (50%)
Frame = +1
Query: 356 PVHLLFFWLTYFLFPVFSLSLLQPLLRVIKMNSVKMQIVGFLFSLISSGFGFYYGHSHWK 415
P+ LLF+++ F + LSLL LL + + N+ M + + +L+ G HWK
Sbjct: 154 PLLLLFYFILSRSFTLCLLSLLLLLLNMSRFNANSMVLFLLILTLV-----LLIGKRHWK 318
>TC78291 weakly similar to PIR|A84793|A84793 nodulin-like protein [imported]
- Arabidopsis thaliana, partial (54%)
Length = 1466
Score = 28.9 bits (63), Expect = 5.2
Identities = 34/132 (25%), Positives = 62/132 (46%), Gaps = 10/132 (7%)
Frame = +1
Query: 229 SQFPIRQGIS-----SKLSLHATAAGCLGSALISAFTYHMIRELNVNEPDIMSLWI---- 279
S F I Q I+ +++SL AT +G+ SA T M R+ P+ SL +
Sbjct: 703 SAFYILQAITLRKYPAEMSL-ATWVCFIGALQSSAVTIFMERK----HPEAWSLGLDSRL 867
Query: 280 -VSIFSGLIWLVGIFHVATAIGRTTDEISFSSRLHPFSVFKYPHAIGALASVFISSFITM 338
S+++G++ F+V ++ +T + F + +P + + ALA + +S + +
Sbjct: 868 FASVYAGIVTSAIQFYVQGSVIKTMGPV-FVTAFNPLRMI----IVTALACIVLSEKLHL 1032
Query: 339 TIFTGGVVFIVG 350
GGVV + G
Sbjct: 1033GSIVGGVVVVTG 1068
>TC84633 similar to PIR|T49099|T49099 dihydrolipoamide S-acetyltransferase
precursor - Arabidopsis thaliana, partial (35%)
Length = 928
Score = 28.5 bits (62), Expect = 6.8
Identities = 18/53 (33%), Positives = 27/53 (49%)
Frame = -1
Query: 287 IWLVGIFHVATAIGRTTDEISFSSRLHPFSVFKYPHAIGALASVFISSFITMT 339
I ++ F VATAI +T+ +S PFS F+Y A G ++ I M+
Sbjct: 307 IGVIKPFSVATAIDMSTESHKMTS---PFSSFQYAFASGTFLRAAATTLIIMS 158
>TC91797 similar to GP|5734493|emb|CAB52700.1 hypothetical protein pucC
homologue {Prochlorococcus marinus}, partial (4%)
Length = 370
Score = 28.5 bits (62), Expect = 6.8
Identities = 17/57 (29%), Positives = 29/57 (50%), Gaps = 5/57 (8%)
Frame = +3
Query: 358 HLLFFWLTYFLFPVFSLSLLQPLLRVIKMNS-----VKMQIVGFLFSLISSGFGFYY 409
+LL ++ Y+ F +F L L I+ S +K ++GF FS+ + F F+Y
Sbjct: 42 YLLTIYILYYFFALFCLIYFSILFYSIQ*KSYYKFRLKPFLIGFFFSISTPIFVFFY 212
>TC81156 similar to PIR|T45754|T45754 hypothetical protein F24M12.270 -
Arabidopsis thaliana, partial (30%)
Length = 1204
Score = 28.1 bits (61), Expect = 8.8
Identities = 10/28 (35%), Positives = 19/28 (67%)
Frame = -2
Query: 345 VVFIVGQLCIKPVHLLFFWLTYFLFPVF 372
++F+ I +HL+FF+++YFL +F
Sbjct: 786 ILFLFFNTLIVIIHLIFFYVSYFLIIIF 703
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.326 0.140 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,851,720
Number of Sequences: 36976
Number of extensions: 302555
Number of successful extensions: 2562
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 2496
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2553
length of query: 535
length of database: 9,014,727
effective HSP length: 101
effective length of query: 434
effective length of database: 5,280,151
effective search space: 2291585534
effective search space used: 2291585534
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0020.10


