

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
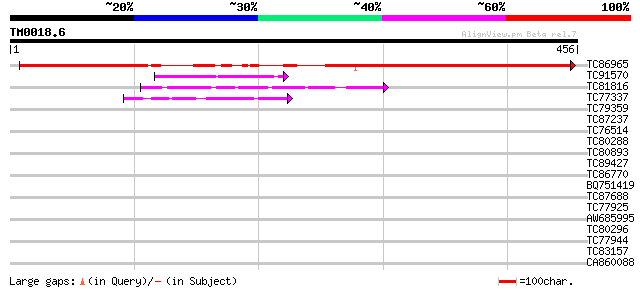
Query= TM0018.6
(456 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86965 similar to GP|22022558|gb|AAM83236.1 At5g12410 {Arabidop... 486 e-138
TC91570 weakly similar to GP|7208782|emb|CAB76913.1 hypothetical... 49 4e-06
TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 47 1e-05
TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Atel... 42 6e-04
TC79359 weakly similar to PIR|S49915|S49915 extensin-like protei... 40 0.001
TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-inf... 40 0.002
TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 39 0.005
TC80288 weakly similar to PIR|T06029|T06029 hypothetical protein... 39 0.005
TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {... 37 0.021
TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed pr... 35 0.046
TC86770 similar to GP|13872875|dbj|BAB43982. contains EST AU0965... 35 0.079
BQ751419 weakly similar to GP|21629340|gb L509.2 {Leishmania maj... 34 0.10
TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~un... 34 0.13
TC77925 similar to GP|18700163|gb|AAL77693.1 AT4g02720/T10P11_1 ... 33 0.23
AW685995 similar to PIR|I51618|I516 nucleolar phosphoprotein - A... 33 0.30
TC80296 similar to PIR|H86265|H86265 protein F3F19.18 [imported]... 32 0.39
TC77944 similar to GP|10179602|gb|AAG13810.1 PSTVd RNA-binding p... 32 0.39
TC83157 similar to GP|21595368|gb|AAM66095.1 unknown {Arabidopsi... 32 0.51
CA860088 similar to GP|8248752|emb hypothetical protein MAL4P2.... 32 0.67
>TC86965 similar to GP|22022558|gb|AAM83236.1 At5g12410 {Arabidopsis
thaliana}, partial (59%)
Length = 1455
Score = 486 bits (1252), Expect = e-138
Identities = 270/449 (60%), Positives = 312/449 (69%), Gaps = 2/449 (0%)
Frame = +1
Query: 9 AAAAGGRKRKRYLPHNKPVKKGSYPLHPGVQGVFITCEGGREHQASREALNILESFYEDL 68
AAAAGG+KRKRYLPHNKPVKKGSYPLHPGVQG FITC+GGREHQA+REALNIL++FYE+L
Sbjct: 76 AAAAGGKKRKRYLPHNKPVKKGSYPLHPGVQGFFITCDGGREHQATREALNILDNFYEEL 255
Query: 69 VDGEHLSVKELLNRPKNKKITFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDS 128
+DG H SV +L ++P NKKITFADSDSSS DDE++EE+ QV EG E DK
Sbjct: 256 IDGAHPSVTKLSDKPLNKKITFADSDSSSIDDDEEEEEKAQVP--EGGEEKEDK------ 411
Query: 129 SVDEHAGIGEKAVGANGAEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGI 188
KPKLD S +N +T + + PK+D G+
Sbjct: 412 -------------------KPKLDVSNSDNTSHDNET----GEKSDPPKID-------GL 501
Query: 189 GEKAEGDEANGDKMPKMDSSVNENAGHDNEIGGKENPHKIDDVHAAAGNKTDDNEGDNDN 248
+A GD+AN DK G ++P I+ +
Sbjct: 502 HAQA-GDKANDDK------------------GDVDSPKTIEKI----------------- 573
Query: 249 IKTKTANELPVVKQCCKTNVPVQDSSNK--VEKSIDTLIDEELRELGDKKKRRFVKLESG 306
A+ELP VK+CCKT P + K EKSID LI++EL EL DK K+RF KLESG
Sbjct: 574 -----ADELPAVKECCKTTAPTSNLGEKKVEEKSIDKLIEDELVELRDKSKKRFAKLESG 738
Query: 307 CNGVVFIQMRKKDGDKSPKDIVHRIVTSAASTRKHMSRFILRMLPIEVSCYASKEEISRA 366
CNGVVFIQMRKKDGDKSP IV+RIVTSAASTRKHMSRFILR+LPIEVSCYASKEEIS+A
Sbjct: 739 CNGVVFIQMRKKDGDKSPNKIVNRIVTSAASTRKHMSRFILRILPIEVSCYASKEEISKA 918
Query: 367 IKPLVEQNFPVPTQKPLKFAVMYEARANTGIDRMEIIDAVAKSVPGPHKVDLKNPDKTIV 426
I+PLVEQ FPV TQ P KFAVMYEARANTG+DRMEIIDAVAKS+P PHKVDL NPDKTI+
Sbjct: 919 IQPLVEQYFPVETQNPQKFAVMYEARANTGVDRMEIIDAVAKSIPAPHKVDLSNPDKTII 1098
Query: 427 VEIARTVCLIGIIEKYKELSKYNLRELTS 455
VEIARTVCLIG++EKYKELSKYN+R+LTS
Sbjct: 1099VEIARTVCLIGVVEKYKELSKYNIRQLTS 1185
>TC91570 weakly similar to GP|7208782|emb|CAB76913.1 hypothetical protein
{Cicer arietinum}, partial (17%)
Length = 1446
Score = 48.9 bits (115), Expect = 4e-06
Identities = 34/109 (31%), Positives = 50/109 (45%), Gaps = 1/109 (0%)
Frame = +2
Query: 117 EANGDK-KLKLDSSVDEHAGIGEKAVGANGAEKPKLDSSVDENAGIGGKTEGDEAKGDEK 175
E N DK K L+ V E +G+K N K L E G+G K + +E G K
Sbjct: 419 EENNDKPKKSLNGGVKEEKDLGQKTQENNDKPKKSLSGGAKEEKGLGQKAQ-EENNGKPK 595
Query: 176 PKLDSSVDEDAGIGEKAEGDEANGDKMPKMDSSVNENAGHDNEIGGKEN 224
L+ S E+ G+ +KA E + D + +S+ + D EIG E+
Sbjct: 596 KSLNGSAKEEKGLRQKALEQEGHNDPNAVLQTSMKKVG--DEEIGVSED 736
>TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum},
partial (13%)
Length = 663
Score = 47.4 bits (111), Expect = 1e-05
Identities = 50/200 (25%), Positives = 88/200 (44%), Gaps = 1/200 (0%)
Frame = +3
Query: 106 EEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGEKAVGANGAEKPKLDSSVDENAGIGGKT 165
EE + + D++ GD K + V+ + K+V + +K K D + +
Sbjct: 69 EESNIPIKIDDKSAGDVK---EDKVEIEKDLEIKSVEKDDEKKEKKDKEKKDKTDVD--- 230
Query: 166 EGDEAKGDEKPKLDSSVDEDAGIGEKAEGDEANGDKMPKMDSSVNENAGHDNEIGGKENP 225
EG + K EK K + E+ GE+ +GDE + K + E D + G+E
Sbjct: 231 EGKDKKDKEKKKKEKK--EENVKGEEEDGDEKKDKEKKKKEKK--EKGKEDKDKDGEEKK 398
Query: 226 HKIDDVHAAAGNKTDDNEGDNDNIKTKTANELPVVKQCCKTNVPVQDSSNKVEKSI-DTL 284
K D N+ DD+EG++ + K K ++ K+ +D + + S+ D
Sbjct: 399 SKKDKEKKKDKNE-DDDEGEDGSKKKKNKDKKEKKKE--------EDEKEEGKVSVRDID 551
Query: 285 IDEELRELGDKKKRRFVKLE 304
I+E +E +KKK++ K E
Sbjct: 552 IEETAKEGKEKKKKKEDKEE 611
Score = 37.4 bits (85), Expect = 0.012
Identities = 44/180 (24%), Positives = 71/180 (39%), Gaps = 4/180 (2%)
Frame = +3
Query: 55 REALNILESFYEDLVDGEHLSVKELLNRPKNKKITFADSDSSSSGDDEDDEEEVQVQVGE 114
+ A ++ E E D E SV++ + + K D G D+ D+E+ + + E
Sbjct: 102 KSAGDVKEDKVEIEKDLEIKSVEKDDEKKEKKDKEKKDKTDVDEGKDKKDKEKKKKEKKE 281
Query: 115 ----GDEANGDKKLKLDSSVDEHAGIGEKAVGANGAEKPKLDSSVDENAGIGGKTEGDEA 170
G+E +GD+K + E G++ +G EK S D+ + DE
Sbjct: 282 ENVKGEEEDGDEKKDKEKKKKEKKEKGKEDKDKDGEEK---KSKKDKEKKKDKNEDDDEG 452
Query: 171 KGDEKPKLDSSVDEDAGIGEKAEGDEANGDKMPKMDSSVNENAGHDNEIGGKENPHKIDD 230
+ K K + E +K E DE K+ D + E A GKE K +D
Sbjct: 453 EDGSKKKKNKDKKE-----KKKEEDEKEEGKVSVRDIDIEETAKE-----GKEKKKKKED 602
>TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Ateline
herpesvirus 3}, partial (8%)
Length = 986
Score = 41.6 bits (96), Expect = 6e-04
Identities = 36/137 (26%), Positives = 58/137 (42%), Gaps = 1/137 (0%)
Frame = +3
Query: 92 DSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGEKAVGANGAEKPKL 151
D++ DDED+E++ +GD A G+ + +L S ++ G G + + ++K
Sbjct: 408 DAEGEDGNDDEDEEDD------DGDGAFGEGEDELSS--EDGGGYGNNSNNKSNSKKAP- 560
Query: 152 DSSVDENAGIGGKTEGDEAKGDEK-PKLDSSVDEDAGIGEKAEGDEANGDKMPKMDSSVN 210
G GG E E + DE D DED ++ EG E D+ +D N
Sbjct: 561 ------EGGAGGADENGEEEDDEDGDDQDEDDDEDEDDDDEEEGGEE--DEEEGVDEEDN 716
Query: 211 ENAGHDNEIGGKENPHK 227
E D + + P K
Sbjct: 717 EEEEEDEDEEALQPPKK 767
>TC79359 weakly similar to PIR|S49915|S49915 extensin-like protein - maize,
partial (3%)
Length = 1170
Score = 40.4 bits (93), Expect = 0.001
Identities = 41/162 (25%), Positives = 64/162 (39%), Gaps = 2/162 (1%)
Frame = -3
Query: 87 KITFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGI-GEKAVGANG 145
K+ S+++G E EE+ +V GD G + V E G+ E+A G +
Sbjct: 736 KLDLEPESSAAAGPCEVSGEELAGEV-PGDLLEGSVAAGASAGVAELEGMPAEEASGGDE 560
Query: 146 AEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIGEKAEGDEANGDKMPKM 205
+ + A +GG+ GDE G E L+ + + G GD A GD
Sbjct: 559 TDGASTGLLTGDTAAVGGELAGDETGGVEDTLLEGLIAGETEDGG*TAGDFAGGDV---- 392
Query: 206 DSSVNENAGHDNEIGGK-ENPHKIDDVHAAAGNKTDDNEGDN 246
+ +A D E G E K+ D AG + E ++
Sbjct: 391 -AGGGVDADPDGETAGVCEGGDKVGDFAGDAGEFVGETETED 269
>TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-infected
erythrocyte surface antigen {Plasmodium falciparum},
partial (2%)
Length = 2007
Score = 39.7 bits (91), Expect = 0.002
Identities = 58/292 (19%), Positives = 114/292 (38%), Gaps = 22/292 (7%)
Frame = +1
Query: 66 EDLVDGEHLSVKELLNRPKNKKITFADSDSSSSGDDEDDEEEVQVQVGEGDE-------- 117
E++ DGE + + N+ + K + ++ + D+E ++E +++ EG E
Sbjct: 172 EEIKDGEKIQQENEENKDEEK----SQQENEENKDEEKSQQENELKKNEGGEKETGEITE 339
Query: 118 ----------ANGDKKLKLDSSVDEHAGIGEKAVGANGAEKPKLDSSVDENAGIGGKTEG 167
+ + K K + +++ ++ VG N + K +E G TEG
Sbjct: 340 EKSKQENEETSETNSKDKENEESNQNGSDAKEQVGENHEQDSK--QGTEETNG----TEG 501
Query: 168 DEAKGDEKPKLDSSVDEDAGIGEKAEGDEANGDKMPKMDSSVNENAGHDNEIGGKENPHK 227
E + +K K D+S D GEK N A +N G +
Sbjct: 502 GEKEEHDKIKEDTSSDNQVQDGEK------------------NNEAREENYSGDNASSAV 627
Query: 228 IDDVHAAAGNKTDDNEGDNDNIKTKTANELPVVKQCCKTNVPVQDSSNKVEKSIDTLIDE 287
+D+ + NKT+ + K NE + Q +SN+ +S D+ I +
Sbjct: 628 VDNKSQESSNKTE------EQFDKKEKNEF---------ELESQKNSNETTESTDSTITQ 762
Query: 288 ELRELGDKKKRRFVKLES----GCNGVVFIQMRKKDGDKSPKDIVHRIVTSA 335
+ G++ ++ + E+ G Q ++++ + + KD V TS+
Sbjct: 763 NSQ--GNESEKDQAQTENDTPKGSASESDEQKQEQEQNNTTKDDVQTTDTSS 912
Score = 33.9 bits (76), Expect = 0.13
Identities = 35/170 (20%), Positives = 67/170 (38%), Gaps = 5/170 (2%)
Frame = +1
Query: 92 DSDSSSSGDDEDDEEEVQV-QVGEGDEANGDKKLKLDSSVDEHAGIGEKAVGANGAEKPK 150
DS S +GD D + +G+ +G+ K + + ++++G ++ +G+
Sbjct: 1015 DSKSGVAGDQADSTTTTSSSETQDGNTNHGEYKDTTNENPEKNSG--QEGTQESGSSSNT 1188
Query: 151 LDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIGEKAEGDEAN-GDKMPKMDSSV 209
D+ + + T D + +K + S+ + E ++ D AN G D S
Sbjct: 1189 FDNKDAASNKVQLTTTSDTSSEQKKDESSSAESKS----ESSQNDNANSGQSNTTSDESA 1356
Query: 210 NENAGHDNEIGGKENPHKIDDVHAAAGNKTDDN---EGDNDNIKTKTANE 256
N+N EN +A GN +N E ND+ + N+
Sbjct: 1357 NDNKDSSQVTTSSEN--------SAEGNSNTENNSDENQNDSKNNENTND 1482
>TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (61%)
Length = 1288
Score = 38.5 bits (88), Expect = 0.005
Identities = 54/256 (21%), Positives = 95/256 (37%), Gaps = 3/256 (1%)
Frame = +3
Query: 86 KKITFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGEKAVGANG 145
KK+ ++ + S DED + + + A K + DE + E+
Sbjct: 510 KKVDTSEEEDSEESSDEDKKPAAKAVPSKNGSAPAKKAASDEEDTDESSDEDEE------ 671
Query: 146 AEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIGEKAEGDEANGDKMPKM 205
EKP + +N + K E+ ++ +SS +ED KA + + K K
Sbjct: 672 DEKPAAKAVPSKNGSVPAKKADTESSDEDS---ESSDEEDKKPAAKASKNVSAPTK--KA 836
Query: 206 DSSVNENAGHDNEIGGKENPHKIDDVHAAAGNKTDDNEGDNDNIKTKTANELPV-VKQCC 264
SS +E + +++ P A K + D D+ + ++ PV K+
Sbjct: 837 ASSSDEESDEESDEDEDAKPVSKPAAVAKKSKKDSSDSDDEDDDSSSDEDKKPVAAKKED 1016
Query: 265 KTNVPV--QDSSNKVEKSIDTLIDEELRELGDKKKRRFVKLESGCNGVVFIQMRKKDGDK 322
K NV DS E+S ++E + KK + +++G K G K
Sbjct: 1017KMNVDKDGSDSDQSEEES-----EDEPSKTPQKKIKDVEMVDAG-----------KSGKK 1148
Query: 323 SPKDIVHRIVTSAAST 338
+P I S + T
Sbjct: 1149APNTPATPIENSGSKT 1196
Score = 36.6 bits (83), Expect = 0.021
Identities = 39/171 (22%), Positives = 75/171 (43%)
Frame = +3
Query: 83 PKNKKITFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGEKAVG 142
P K + + SS +DE+DE+ V + + KK +SS DE + ++
Sbjct: 609 PAKKAASDEEDTDESSDEDEEDEKPAAKAVPSKNGSVPAKKADTESS-DEDSESSDEEDK 785
Query: 143 ANGAEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIGEKAEGDEANGDKM 202
A+ K S+ + A E DE +++ V + A + +K++ D ++ D
Sbjct: 786 KPAAKASKNVSAPTKKAASSSDEESDEESDEDEDA--KPVSKPAAVAKKSKKDSSDSDDE 959
Query: 203 PKMDSSVNENAGHDNEIGGKENPHKIDDVHAAAGNKTDDNEGDNDNIKTKT 253
DSS +E+ KE+ +D G+ +D +E ++++ +KT
Sbjct: 960 DD-DSSSDED--KKPVAAKKEDKMNVD----KDGSDSDQSEEESEDEPSKT 1091
>TC80288 weakly similar to PIR|T06029|T06029 hypothetical protein T28I19.100
- Arabidopsis thaliana, partial (14%)
Length = 1460
Score = 38.5 bits (88), Expect = 0.005
Identities = 64/302 (21%), Positives = 109/302 (35%), Gaps = 40/302 (13%)
Frame = +3
Query: 27 VKKGSYPLHPGVQGVFITCEGGREHQASREALNILESFYEDLVDGEHLSVKELLNRPKNK 86
+K G LHPG + + H+ E +I+ + E E N+ + +
Sbjct: 339 LKLGRKDLHPGK----VEADKNEGHEEEEEDEHIVYNMQNKREHDEQQQEGEEGNKHETE 506
Query: 87 KITFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGEKAVGANGA 146
+ D+ +E DEEE + G E + + K + DE G+ + N
Sbjct: 507 E---ESEDNVHERREEQDEEENK----HGAEVQEENESKSEEVEDEG---GDVEIDENDH 656
Query: 147 EKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDED-AGIGEKAEGDEANGDKMPKM 205
EK + D+ D + + + E +GD++ + + DE+ G+ E E EA +
Sbjct: 657 EKSEADN--DREDEVVDEEKDKEEEGDDETENEDKEDEEKGGLVENHENHEAREEHYKAD 830
Query: 206 DSS-----------------------------VNENAGHDNEIGGKENPHKI-------- 228
D+S H +E G +N +
Sbjct: 831 DASSAVAHDTHETSTETGNLEHSDLSLQNTRKPENETNHSDESYGSQNVSDLKVTEGELT 1010
Query: 229 DDV--HAAAGNKTDDNEGDNDNIKTKTANELPVVKQCCKTNVPVQDSSNKVEKSIDTLID 286
D V +A AG +T ++ N+ KTK ++L + T V + SSN DT
Sbjct: 1011DGVSSNATAGKETGNDSFSNETAKTKPDSQLDLSSNL--TAVITEASSNSSGTGDDTSSS 1184
Query: 287 EE 288
E
Sbjct: 1185SE 1190
>TC80893 similar to GP|4557063|gb|AAD22502.1| expressed protein {Arabidopsis
thaliana}, partial (39%)
Length = 883
Score = 36.6 bits (83), Expect = 0.021
Identities = 32/122 (26%), Positives = 54/122 (44%), Gaps = 3/122 (2%)
Frame = +3
Query: 86 KKITFADSDSSSSGD-DEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGEKAVGAN 144
+K + ++ S + D D+DD+++VQ + +G+E E G
Sbjct: 288 RKPAYKENKSDTEDDEDDDDDDDVQDEDDDGEE--------------------EDYSGDE 407
Query: 145 GAEKPKLDSSVDENAGIGGKTEG--DEAKGDEKPKLDSSVDEDAGIGEKAEGDEANGDKM 202
G E+ + + N G GG +G D+ GDE+ DE+ G E+ E DE D+
Sbjct: 408 GEEEGDPEDDPEAN-GAGGSDDGEDDDDDGDEE-------DEEDGEDEEDEEDEEEDDET 563
Query: 203 PK 204
P+
Sbjct: 564 PQ 569
Score = 28.1 bits (61), Expect = 7.4
Identities = 35/126 (27%), Positives = 48/126 (37%), Gaps = 3/126 (2%)
Frame = +3
Query: 126 LDSSVDEHAGIGEKAVGANGAEKPKL---DSSVDENAGIGGKTEGDEAKGDEKPKLDSSV 182
L S+ A IG + A + PKL DS +G K E K D + D
Sbjct: 168 LVSAAQSLALIGYLLINAANSVAPKLTAMDSRFPLEHLLGRKPAYKENKSDTEDDEDDDD 347
Query: 183 DEDAGIGEKAEGDEANGDKMPKMDSSVNENAGHDNEIGGKENPHKIDDVHAAAGNKTDDN 242
D+D E +G+E D S +E G+E DD A +DD
Sbjct: 348 DDDVQ-DEDDDGEEE--------DYSGDE---------GEEEGDPEDDPEANGAGGSDDG 473
Query: 243 EGDNDN 248
E D+D+
Sbjct: 474 EDDDDD 491
>TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed protein
{Arabidopsis thaliana}, partial (50%)
Length = 753
Score = 35.4 bits (80), Expect = 0.046
Identities = 26/86 (30%), Positives = 40/86 (46%)
Frame = +1
Query: 162 GGKTEGDEAKGDEKPKLDSSVDEDAGIGEKAEGDEANGDKMPKMDSSVNENAGHDNEIGG 221
G +TE D+ + D+ D DED E GDE + D P+ D N G D++
Sbjct: 298 GSETEDDDDEDDD----DDVNDEDDDNDEDFSGDEDDEDADPEDDPVPNGAGGSDDD--- 456
Query: 222 KENPHKIDDVHAAAGNKTDDNEGDND 247
E+ DD + ++ +D E D+D
Sbjct: 457 DEDDDDDDDDNDDGEDEDEDEEEDDD 534
Score = 32.3 bits (72), Expect = 0.39
Identities = 23/86 (26%), Positives = 41/86 (46%), Gaps = 3/86 (3%)
Frame = +1
Query: 92 DSDSSSSGDDEDDEEEVQVQVGEGDE-ANGDKKLKLDSSVDEHAGIGEKAV--GANGAEK 148
D + DDEDD+++V + + DE +GD+ DE A + V GA G++
Sbjct: 295 DGSETEDDDDEDDDDDVNDEDDDNDEDFSGDED-------DEDADPEDDPVPNGAGGSDD 453
Query: 149 PKLDSSVDENAGIGGKTEGDEAKGDE 174
D D++ G+ E ++ + D+
Sbjct: 454 DDEDDDDDDDDNDDGEDEDEDEEEDD 531
Score = 30.4 bits (67), Expect = 1.5
Identities = 20/64 (31%), Positives = 28/64 (43%), Gaps = 5/64 (7%)
Frame = +1
Query: 190 EKAEGDEANGDKMPKMDSSVN-ENAGHDNEIGGKENPHKIDD----VHAAAGNKTDDNEG 244
E +G E D D VN E+ +D + G E+ D V AG DD+E
Sbjct: 286 ENKDGSETEDDDDEDDDDDVNDEDDDNDEDFSGDEDDEDADPEDDPVPNGAGGSDDDDED 465
Query: 245 DNDN 248
D+D+
Sbjct: 466 DDDD 477
>TC86770 similar to GP|13872875|dbj|BAB43982. contains EST
AU096506(S14064)~unknown protein {Oryza sativa (japonica
cultivar-group)}, partial (73%)
Length = 2061
Score = 34.7 bits (78), Expect = 0.079
Identities = 38/135 (28%), Positives = 57/135 (42%), Gaps = 15/135 (11%)
Frame = +2
Query: 179 DSSVDEDAGIGEKAEGDEANGDKMPKMDSSVNENA-------GHDNEIGGKENPHKIDDV 231
D +ED G K D +G K ++ + + G +GGK + +V
Sbjct: 374 DELEEEDEGRKRKKGKDGGSGKKEKRLKGGSSSSGKGGSRFGGSKRGVGGKSGNDREGEV 553
Query: 232 H----AAAGNKTDDNEG----DNDNIKTKTANELPVVKQCCKTNVPVQDSSNKVEKSIDT 283
+ A AGN DDNEG D+DN T E P + + P D+ E D
Sbjct: 554 NEMWDALAGNSEDDNEGARNMDDDNFIDDTGVE-PALYGYDEPRSP-GDAPQAEEGEEDD 727
Query: 284 LIDEELRELGDKKKR 298
I ++L ++G KKK+
Sbjct: 728 EI-KDLFKMGKKKKK 769
>BQ751419 weakly similar to GP|21629340|gb L509.2 {Leishmania major}, partial
(1%)
Length = 766
Score = 34.3 bits (77), Expect = 0.10
Identities = 25/63 (39%), Positives = 32/63 (50%), Gaps = 5/63 (7%)
Frame = -3
Query: 140 AVGANGAEKPKLDSSVDENAGIGGKTE----GDEAKGDEKPKLDSSVDEDAG-IGEKAEG 194
A+ G E LD+ E AG GG E GD +GD LD+ VDED G + + EG
Sbjct: 407 ALARGGREVELLDA---EGAGAGGVPELWGEGDVVRGDVAAGLDAEVDEDVGPVVVQVEG 237
Query: 195 DEA 197
+ A
Sbjct: 236 EGA 228
>TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~unknown
protein {Arabidopsis thaliana}, partial (46%)
Length = 2077
Score = 33.9 bits (76), Expect = 0.13
Identities = 41/171 (23%), Positives = 64/171 (36%), Gaps = 6/171 (3%)
Frame = +2
Query: 92 DSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGEKAVGANGAEKPKL 151
D + +DEDDEE+ + +V G GI +K +
Sbjct: 494 DDEDEEGSEDEDDEEDEKEKVKGG-------------------GIEDKFL---------- 586
Query: 152 DSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDEDAGIGEKA---EGDEANGDKMPKMDSS 208
+DE K E + KG+E+ + D +ED + EKA E D+ + D + D
Sbjct: 587 --KIDELTEYLEKEEDNYEKGEERDEADEDSEEDDEL-EKAGEFEMDDEDDDDDDEEDEE 757
Query: 209 VNE--NAGHDNEIGGK-ENPHKIDDVHAAAGNKTDDNEGDNDNIKTKTANE 256
+ N +++ GGK E K D DD E +T + E
Sbjct: 758 AEDMGNVRYEDFFGGKNEKGSKRKDQLLEVSGDEDDMESTKQKKRTASTYE 910
>TC77925 similar to GP|18700163|gb|AAL77693.1 AT4g02720/T10P11_1
{Arabidopsis thaliana}, partial (47%)
Length = 1538
Score = 33.1 bits (74), Expect = 0.23
Identities = 41/183 (22%), Positives = 73/183 (39%), Gaps = 7/183 (3%)
Frame = +3
Query: 70 DGEHLSVKELLNRPKNKKITFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKL--- 126
D E S E+ +R + K +++SDS ++ ++ED + + + + KK +
Sbjct: 633 DSESESESEVEDRKRRKSRKYSESDSDTNSEEEDRKRRRRRKSSRRRRKSSRKKRRCSDS 812
Query: 127 -DSSVDEHAGI---GEKAVGANGAEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSV 182
+S D+ +G G K + + K K SS + G G ++ + + V
Sbjct: 813 DESETDDESGYDDSGRKRKRSKRSRKSKKKSSEPVSEGSEEIELGSDSAVAKINEEIDDV 992
Query: 183 DEDAGIGEKAEGDEANGDKMPKMDSSVNENAGHDNEIGGKENPHKIDDVHAAAGNKTDDN 242
DE EK A K+ ++ S + A D+E P + H + G
Sbjct: 993 DE-----EKMAEMNAEALKLKELFESQKKPALDDDEPPVGPMPLPRAEGHISYGGALRPG 1157
Query: 243 EGD 245
EGD
Sbjct: 1158EGD 1166
>AW685995 similar to PIR|I51618|I516 nucleolar phosphoprotein - African
clawed frog, partial (5%)
Length = 516
Score = 32.7 bits (73), Expect = 0.30
Identities = 29/102 (28%), Positives = 48/102 (46%), Gaps = 8/102 (7%)
Frame = +3
Query: 105 EEEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGE------KAVGANGAEKPKLDSSVDEN 158
EEE + Q + +E +++ DSS E + E KA A A K SS +E+
Sbjct: 117 EEEKKKQQKKKEEKPAPMEVEEDSSSSEESSSSEDEAPAKKAAPAKKAAAKKESSSEEES 296
Query: 159 AGIGGKTEGDE--AKGDEKPKLDSSVDEDAGIGEKAEGDEAN 198
+ ++E +E AK K SS +E + E+++ +E N
Sbjct: 297 SSSESESEEEEKPAKKAAAKKESSSEEESSSSEEESDEEEKN 422
>TC80296 similar to PIR|H86265|H86265 protein F3F19.18 [imported] -
Arabidopsis thaliana, partial (34%)
Length = 1734
Score = 32.3 bits (72), Expect = 0.39
Identities = 29/98 (29%), Positives = 42/98 (42%), Gaps = 2/98 (2%)
Frame = +3
Query: 100 DDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHAGIGEKAVGANGAEKPKLDSSVDENA 159
DD+ ++E D A D ++ L+S D G + AE S + +
Sbjct: 756 DDDVEQESSHSDDCGSDNAQEDDQVSLNSDDDNQLGSDNTGSDDDEAEDHDGVSDDENDR 935
Query: 160 GIGGKTEGDEAKG--DEKPKLDSSVDEDAGIGEKAEGD 195
+T GD+A DE L+ S +ED GI E EGD
Sbjct: 936 SSDYETSGDDADNVEDEGDDLEDS-EEDGGISEH-EGD 1043
Score = 27.7 bits (60), Expect = 9.6
Identities = 16/66 (24%), Positives = 29/66 (43%)
Frame = +3
Query: 182 VDEDAGIGEKAEGDEANGDKMPKMDSSVNENAGHDNEIGGKENPHKIDDVHAAAGNKTDD 241
+D+D E + D+ D + D V+ N+ DN++G D+ G D+
Sbjct: 753 IDDDVE-QESSHSDDCGSDNAQE-DDQVSLNSDDDNQLGSDNTGSDDDEAEDHDGVSDDE 926
Query: 242 NEGDND 247
N+ +D
Sbjct: 927 NDRSSD 944
>TC77944 similar to GP|10179602|gb|AAG13810.1 PSTVd RNA-binding protein
Virp1a {Lycopersicon esculentum}, partial (27%)
Length = 1144
Score = 32.3 bits (72), Expect = 0.39
Identities = 21/83 (25%), Positives = 38/83 (45%), Gaps = 5/83 (6%)
Frame = +1
Query: 143 ANGAEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDED-----AGIGEKAEGDEA 197
+ G ++ K+D+ VDE+ IG D+ + P ++ D+D G + G +
Sbjct: 403 SEGKKQKKIDT-VDEDVDIG-----DDMPANNFPPVEIEKDKDMGATGGGASSSSSGSSS 564
Query: 198 NGDKMPKMDSSVNENAGHDNEIG 220
+G DS ++G D+E G
Sbjct: 565 SGSDSSSSDSDSGSSSGSDSEAG 633
>TC83157 similar to GP|21595368|gb|AAM66095.1 unknown {Arabidopsis
thaliana}, partial (57%)
Length = 1108
Score = 32.0 bits (71), Expect = 0.51
Identities = 18/59 (30%), Positives = 30/59 (50%), Gaps = 8/59 (13%)
Frame = +3
Query: 92 DSDSSSSGDDEDDEEEVQVQVGEGDEANGD-------KKLKLDSSVDEHA-GIGEKAVG 142
D D S GDD+DD+++ + G+ ++ + D KK+ + + A IG K VG
Sbjct: 342 DEDDDSDGDDDDDDDDYDEEEGDYEDEDDDTVAVSSRKKVYTEEEKEAEAEAIGYKVVG 518
>CA860088 similar to GP|8248752|emb hypothetical protein MAL4P2.29
{Plasmodium falciparum 3D7}, partial (1%)
Length = 455
Score = 31.6 bits (70), Expect = 0.67
Identities = 20/58 (34%), Positives = 27/58 (46%)
Frame = +1
Query: 206 DSSVNENAGHDNEIGGKENPHKIDDVHAAAGNKTDDNEGDNDNIKTKTANELPVVKQC 263
+SS N N+ + N I N KI D + N DDN+ +N K + NE K C
Sbjct: 154 NSSENNNSINSNNIENIPNIPKILD----SNNNDDDNDNNNKESKIENTNEHNFQKNC 315
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.308 0.131 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,290,824
Number of Sequences: 36976
Number of extensions: 140117
Number of successful extensions: 1078
Number of sequences better than 10.0: 78
Number of HSP's better than 10.0 without gapping: 895
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 991
length of query: 456
length of database: 9,014,727
effective HSP length: 99
effective length of query: 357
effective length of database: 5,354,103
effective search space: 1911414771
effective search space used: 1911414771
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0018.6


