

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0017.9
(387 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
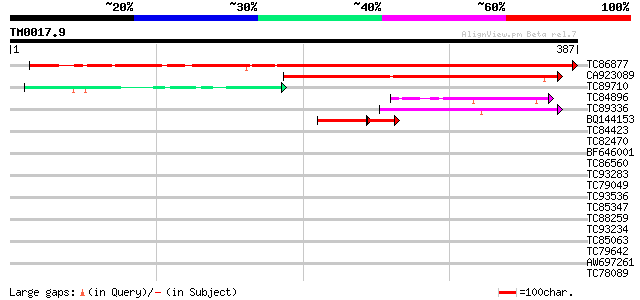
Score E
Sequences producing significant alignments: (bits) Value
TC86877 similar to GP|8953376|emb|CAB96649.1 putative protein {A... 376 e-105
CA923089 homologue to GP|3114816|emb| JM1 {Homo sapiens}, partia... 240 8e-64
TC89710 homologue to GP|13660721|gb|AAK32956.1 cytochrome P450 {... 75 4e-14
TC84896 similar to GP|7596769|gb|AAF64540.1| unknown protein {Ar... 44 8e-05
TC89336 weakly similar to GP|21554135|gb|AAM63215.1 unknown {Ara... 44 1e-04
BQ144153 similar to GP|8953376|emb| putative protein {Arabidopsi... 31 4e-04
TC84423 similar to PIR|F84730|F84730 probable myosin heavy chain... 39 0.004
TC82470 homologue to GP|23497460|gb|AAN37003.1 hypothetical prot... 38 0.008
BF646001 homologue to GP|20303561|gb| putative kinase {Oryza sat... 36 0.029
TC86560 similar to GP|8843737|dbj|BAA97285.1 myosin heavy chain-... 36 0.029
TC93283 weakly similar to GP|23476966|emb|CAD43403. SMC3 protein... 35 0.038
TC79049 weakly similar to PIR|T10798|T10798 pherophorin-S - Volv... 35 0.064
TC93536 weakly similar to GP|2961371|emb|CAA18118.1 hypothetical... 35 0.064
TC85347 similar to GP|22136328|gb|AAM91242.1 unknown protein {Ar... 35 0.064
TC88259 similar to GP|6729037|gb|AAF27033.1| unknown protein {Ar... 34 0.11
TC93234 weakly similar to GP|21751020|dbj|BAC03887. unnamed prot... 33 0.24
TC85063 similar to PIR|T47959|T47959 hypothetical protein F15G16... 33 0.24
TC79642 similar to PIR|T07397|T07397 kinesin heavy chain-like pr... 33 0.24
AW697261 similar to GP|14596027|gb| Unknown protein {Arabidopsis... 33 0.24
TC78089 similar to GP|4522009|gb|AAD21782.1| unknown protein {Ar... 32 0.32
>TC86877 similar to GP|8953376|emb|CAB96649.1 putative protein {Arabidopsis
thaliana}, partial (12%)
Length = 1558
Score = 376 bits (965), Expect = e-105
Identities = 215/383 (56%), Positives = 280/383 (72%), Gaps = 9/383 (2%)
Frame = +2
Query: 14 CPHASNPYHQCTQACSKKTSHRTKPHAKNTSSSGAAPPPSNFDRRKKPGSRPEPPVVAGA 73
CP ASNPYH C C K+ S + SG P FDR+KK GSRPE PV+ +
Sbjct: 356 CPKASNPYHACDVNCQKRMSG---------ADSGVIP--LTFDRKKKLGSRPELPVL-DS 499
Query: 74 VPAAKVEAIQKSNASSPISNHSEIKKVESRNDDHNHSVPVSGQIHVPLPVPDVKHVHEED 133
VPA+K+ AI ++A+SPIS + + KK+E ++ N VP SG++H DVK V+++
Sbjct: 500 VPASKIGAIYLADAASPISKYYDKKKLEPKS---NEIVPASGELHTL----DVKPVNDKV 658
Query: 134 QQKNGVEHLANPIQTKQE-DKTASD-VHP-------GEGLATTFKGGSIDFSFSGIIPQG 184
Q K+G E+LA IQT Q DK +S+ V P GEGL TT GGS F +S ++
Sbjct: 659 QPKDGSENLAGQIQTNQAGDKNSSNKVVPVTCFDDIGEGL-TTSAGGSKHFCYSDVL-HD 832
Query: 185 NEDNDDKGDTESVVSESRVPVGKYHVKESFASILESILDKYGDIGASCHLESLVLRSYYI 244
NED+DD+ +ESVVSE+RVPVGKYHVKESFA IL+SI DKYGDIG SCHLES+V+RSYYI
Sbjct: 833 NEDSDDEEGSESVVSETRVPVGKYHVKESFAPILQSIFDKYGDIGESCHLESVVMRSYYI 1012
Query: 245 ECVCFVVQELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQH 304
ECVCFV+QELQS SIM LTKSK+ EL+AIL DVESAQL V WLR+ ++EIAE +++I++H
Sbjct: 1013ECVCFVIQELQSTSIMDLTKSKVKELLAILNDVESAQLHVTWLRTVVNEIAEYIKLIDEH 1192
Query: 305 QEVDAAKANTDREMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELE 364
+ V+AAKAN+D EMESLR++LES++E L +KE+EV DIK +I I +RL ELE+KS++LE
Sbjct: 1193RSVEAAKANSDSEMESLRKELESKVEILTQKEREVTDIKTKIEGIRERLGELEMKSSDLE 1372
Query: 365 QSMLSSKSEVENLHRKSLIDELL 387
++ LS KS+V+NL +SL+DELL
Sbjct: 1373KNRLSIKSKVDNLDSRSLLDELL 1441
>CA923089 homologue to GP|3114816|emb| JM1 {Homo sapiens}, partial (2%)
Length = 793
Score = 240 bits (612), Expect = 8e-64
Identities = 127/193 (65%), Positives = 161/193 (82%), Gaps = 3/193 (1%)
Frame = -2
Query: 188 NDDKGDTESVVSESRVPVGKYHVKESFASILESILDKYGDIGASCHLESLVLRSYYIECV 247
++ + D+ SVVSESRV +GKY+VKESF SIL++I+DKYGDIGASC LES+V+RSYY+ECV
Sbjct: 771 SEGEADSVSVVSESRVSIGKYNVKESFGSILQTIVDKYGDIGASCDLESVVMRSYYMECV 592
Query: 248 CFVVQELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEV 307
CFVVQELQS S ++KSK++EL+ I+KDVESA LRVAWL + LDEI EN+E+I+ HQ++
Sbjct: 591 CFVVQELQSSSDS-ISKSKVSELLDIVKDVESAHLRVAWLHNTLDEIVENIELISHHQDM 415
Query: 308 DAAKANTDREMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELE-LKSAEL--E 364
+ KAN DREMESLREQLESE+E+LA+KEQEVADI RIPEI DRL ELE L S+ L +
Sbjct: 414 EMEKANYDREMESLREQLESELETLAQKEQEVADINIRIPEIRDRLSELEQLMSSGLVDD 235
Query: 365 QSMLSSKSEVENL 377
Q+ L KS+++ L
Sbjct: 234 QTTLPIKSKIDQL 196
>TC89710 homologue to GP|13660721|gb|AAK32956.1 cytochrome P450 {Anopheles
gambiae}, partial (2%)
Length = 619
Score = 75.1 bits (183), Expect = 4e-14
Identities = 64/222 (28%), Positives = 86/222 (37%), Gaps = 43/222 (19%)
Frame = -2
Query: 11 NPNCPHASNPYHQCTQACSKKTSHRTKPHAKN------------------------TSSS 46
N +C +ASNPYHQCTQACS+KT HA SSS
Sbjct: 561 NGSCANASNPYHQCTQACSQKTKGTKTHHAPTAAVTASNSNSNNRKVINGGERRTYASSS 382
Query: 47 GAAP-------------------PPSNFDRRKKPGSRPEPPVVAGAVPAAKVEAIQKSNA 87
+ P P S D RKK GS+P+PPV+ P
Sbjct: 381 SSCPKSSNPYHKCDANCNNSGATPHSKIDHRKKVGSKPQPPVLHSVPPT----------- 235
Query: 88 SSPISNHSEIKKVESRNDDHNHSVPVSGQIHVPLPVPDVKHVHEEDQQKNGVEHLANPIQ 147
K V ++ND+ +P SG I L +PDV +DQ K+G
Sbjct: 234 ----------KLVATKNDE---IIPTSGPISAQLHIPDVM---PKDQVKDGA-------- 127
Query: 148 TKQEDKTASDVHPGEGLATTFKGGSIDFSFSGIIPQGNEDND 189
T+ + + ++ P T + GS DFSFSG P + +
Sbjct: 126 TEVKVAASHEIVPVTNSNETHEXGSKDFSFSGNPPSSPQQRN 1
>TC84896 similar to GP|7596769|gb|AAF64540.1| unknown protein {Arabidopsis
thaliana}, partial (16%)
Length = 786
Score = 44.3 bits (103), Expect = 8e-05
Identities = 38/117 (32%), Positives = 64/117 (54%), Gaps = 6/117 (5%)
Frame = +1
Query: 261 HLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEV-DAAKANTD--RE 317
+ TKSK +L+++LK L S+L + E + + QE+ DA +N+D +E
Sbjct: 124 YTTKSK-GDLISVLKAEREE------LESSLSK--EKLHTLQLKQELADAESSNSDLYKE 276
Query: 318 MESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELEL---KSAELEQSMLSSK 371
++S+R QL +E + E EVA++ +++ KELEL + A EQ+ LS+K
Sbjct: 277 LQSVRGQLAAEQSRCFKLEVEVAELGQKLQNFGTLQKELELLQRQKAASEQAALSAK 447
>TC89336 weakly similar to GP|21554135|gb|AAM63215.1 unknown {Arabidopsis
thaliana}, partial (7%)
Length = 1241
Score = 43.9 bits (102), Expect = 1e-04
Identities = 34/133 (25%), Positives = 64/133 (47%), Gaps = 8/133 (6%)
Frame = +1
Query: 253 ELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEVDAAK- 311
E Q DS+ ++EL V Q + A L + + + E +EV N+H ++ +
Sbjct: 580 EGQLDSVQKELTLNMDELEHKKGQVLELQKQKAELETHVPNLVEQLEVANEHLKISNDEV 759
Query: 312 ANTDREMES-------LREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELE 364
A +E+ES L++QLE E++ + E ++ +K+I E+ DR+ + LE
Sbjct: 760 ARLRKELESNLAETRQLQDQLEVAQENVTKLEWQLDSGRKQIRELEDRITWFKTNETNLE 939
Query: 365 QSMLSSKSEVENL 377
+ K E+ ++
Sbjct: 940 VEVQKLKDEMHDV 978
>BQ144153 similar to GP|8953376|emb| putative protein {Arabidopsis thaliana},
partial (5%)
Length = 885
Score = 30.8 bits (68), Expect(2) = 4e-04
Identities = 19/39 (48%), Positives = 26/39 (65%), Gaps = 2/39 (5%)
Frame = +3
Query: 211 KESFASILESILDKYGDIGASC-HLESLVLRSYYIE-CV 247
+ESFA +L+SI DKYG I + + L + SYYI+ CV
Sbjct: 6 RESFAPMLQSIFDKYGHITRNLPFVICLRMLSYYIKVCV 122
Score = 30.4 bits (67), Expect(2) = 4e-04
Identities = 13/22 (59%), Positives = 17/22 (77%)
Frame = +1
Query: 245 ECVCFVVQELQSDSIMHLTKSK 266
+CVCFV+QE +S SIM L K +
Sbjct: 112 KCVCFVIQE*KSTSIMDLQKCR 177
>TC84423 similar to PIR|F84730|F84730 probable myosin heavy chain [imported]
- Arabidopsis thaliana, partial (10%)
Length = 787
Score = 38.5 bits (88), Expect = 0.004
Identities = 46/185 (24%), Positives = 79/185 (41%), Gaps = 15/185 (8%)
Frame = +2
Query: 218 LESILDKYGDIGASCHLESLVLRSYYIECVCFVVQELQSDSIMHLTKSKINELVAILKDV 277
LE + ++ G + A+ SL L + IE +E +S L + + + A K+V
Sbjct: 221 LEDLHNESGAVAATASQRSLELEGH-IEATNAAAEEAKSQ----LRELETRFIAAEQKNV 385
Query: 278 E-SAQLRVAWLRSALDEIAENVEVINQHQEVDAAKANTDREMESLREQLESEMESLAEKE 336
E QL + L+ A D + E + +DA + E L L+ M+ L++ E
Sbjct: 386 ELEQQLNLVQLK-ANDAERDVTEFSEKISHLDAKLKEAEEEKNLLNSLLQEHMDKLSQLE 562
Query: 337 --------------QEVADIKKRIPEISDRLKELELKSAELEQSMLSSKSEVENLHRKSL 382
+E+ +K++ E DR +S ELE + SS S+ E +++
Sbjct: 563 SDLNQSTQKNSQLEEELKIVKEKCSEHEDRATMNNERSRELEDLIQSSHSKSEKCEKRAS 742
Query: 383 IDELL 387
ELL
Sbjct: 743 ELELL 757
>TC82470 homologue to GP|23497460|gb|AAN37003.1 hypothetical protein
{Plasmodium falciparum 3D7}, partial (8%)
Length = 1064
Score = 37.7 bits (86), Expect = 0.008
Identities = 44/220 (20%), Positives = 97/220 (44%), Gaps = 21/220 (9%)
Frame = +2
Query: 182 PQGNEDNDDKGDTESVVSESRVPVGKYHVKESFASILESIL---DKYGDIGASCHLESLV 238
P ++D+DD D + + + K SF+++ E ++ + DI + +E
Sbjct: 29 PYDDDDDDDDDDDDDDDDDDDMSTDK-----SFSTLKEKLVLVEKSFEDIRSKTQVEERR 193
Query: 239 LRSYY--IECVCFVVQELQSD-----SIMHLTKSKINELVAILKDVESAQLRVAWLRSAL 291
L+S IE C ++ + + I+ K ++ +KD + + ++ + +
Sbjct: 194 LQSIKRDIEECCEDLENKKKEIRDVGRIIEARKKMQGKIDECVKDFVAKEGQLGLMEDLI 373
Query: 292 DE-----------IAENVEVINQHQEVDAAKANTDREMESLREQLESEMESLAEKEQEVA 340
E + + ++ I++ +E+++ ++ S ++ ES ++ L KE++
Sbjct: 374 GEHKKELKTKELELRQVMDNISKQKELESQVKELVNDLVSKQKHFESHIKELESKERQ-- 547
Query: 341 DIKKRIPEISDRLKELELKSAELEQSMLSSKSEVENLHRK 380
++ R+ E KE E + ELE KSEVE ++ K
Sbjct: 548 -LEGRLKEHELEEKEFEGRMNELESKERHFKSEVEEINAK 664
>BF646001 homologue to GP|20303561|gb| putative kinase {Oryza sativa
(japonica cultivar-group)}, partial (6%)
Length = 667
Score = 35.8 bits (81), Expect = 0.029
Identities = 25/110 (22%), Positives = 50/110 (44%), Gaps = 15/110 (13%)
Frame = +2
Query: 292 DEIAENVEVINQHQEVDAAKANTD-------------REMESLREQLESEMESLAEKEQE 338
+ I N E+ E++ + N+D RE+ + ++E + + EKE +
Sbjct: 191 ENIKNNEEIEKMKTEIEKLRKNSDSTAELEKEAARLRREVVESKVEIEKLRKIIDEKENK 370
Query: 339 VADIKKRIPEISDRLKELELKSAELEQ--SMLSSKSEVENLHRKSLIDEL 386
+ ++K E+ E+E+K ELE+ ++ K EN R + +E+
Sbjct: 371 IEIVEKEGKELKQENVEMEMKVRELERRIGVIEMKEVEENSKRVRIEEEM 520
Score = 33.9 bits (76), Expect = 0.11
Identities = 26/79 (32%), Positives = 45/79 (56%), Gaps = 4/79 (5%)
Frame = +2
Query: 311 KANTDREMESLR-EQLESEMESLAEKEQEVADIKKRIPEI---SDRLKELELKSAELEQS 366
K ++++ M L E+ E E E++ E E+ +K I ++ SD ELE ++A L +
Sbjct: 131 KGSSEKIMAVLECERGELETENIKNNE-EIEKMKTEIEKLRKNSDSTAELEKEAARLRRE 307
Query: 367 MLSSKSEVENLHRKSLIDE 385
++ SK E+E L + +IDE
Sbjct: 308 VVESKVEIEKL--RKIIDE 358
>TC86560 similar to GP|8843737|dbj|BAA97285.1 myosin heavy chain-like
{Arabidopsis thaliana}, partial (28%)
Length = 2450
Score = 35.8 bits (81), Expect = 0.029
Identities = 38/124 (30%), Positives = 66/124 (52%), Gaps = 10/124 (8%)
Frame = +2
Query: 262 LTKSKINELVAILKDV--ESAQLRVAWLRSALDEIAE-NVEVINQHQ-EVDAAKANT--- 314
L +SK +L+A+ K+ E + A L ++EIA + E+ N+ ++D+ K T
Sbjct: 1049 LEQSK-KKLLALKKEFNPEITKSLEAQLAETMNEIAALHTELENKRSSDLDSVKTVTSEL 1225
Query: 315 DREMESLREQLESE--MESLAEK-EQEVADIKKRIPEISDRLKELELKSAELEQSMLSSK 371
D ESL++ ++ E + SL E + E+ ++KK E+ ++ ELE L + SK
Sbjct: 1226 DGAKESLQKVVDEENTLRSLVETLKVELENVKKEHSELKEKESELESTVGNLHVKLRKSK 1405
Query: 372 SEVE 375
SE+E
Sbjct: 1406 SELE 1417
>TC93283 weakly similar to GP|23476966|emb|CAD43403. SMC3 protein
{Arabidopsis thaliana}, partial (18%)
Length = 902
Score = 35.4 bits (80), Expect = 0.038
Identities = 16/55 (29%), Positives = 31/55 (56%)
Frame = +1
Query: 325 LESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSKSEVENLHR 379
++ +S+ KE+E+ +K I EI ++ +L + +++ SKSE+E L R
Sbjct: 391 IKQNADSIHVKEEELEKVKFEIQEIDQKINDLVTEQQKVDAQCAHSKSEIEELKR 555
Score = 35.4 bits (80), Expect = 0.038
Identities = 37/169 (21%), Positives = 79/169 (45%), Gaps = 28/169 (16%)
Frame = +1
Query: 235 ESLVLRSYYIECVCFVVQELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEI 294
+S+ ++ +E V F +QE+ KIN+LV + V++ + A +S ++E+
Sbjct: 406 DSIHVKEEELEKVKFEIQEIDQ---------KINDLVTEQQKVDA---QCAHSKSEIEEL 549
Query: 295 AENVEVINQHQEV-DAAKANTDREMESLREQLESEMESLAEKEQEV-------------- 339
++ N+ + + A A ++ +E ++ Q+E S+A K+ E+
Sbjct: 550 KRDIANSNKQKPLFSKALAKKEKSLEDVQNQIEQLKGSIATKKAEMGTELIDHLTPXEKK 729
Query: 340 --ADIKKRIPEISDRLK-------ELELKSAELEQSMLSS----KSEVE 375
+D+ I ++ ++L E E + AELE ++ ++ K E+E
Sbjct: 730 LLSDLNPEIKDLKEKLVACKTDRIESEARKAELETNLTTNLXKXKQELE 876
>TC79049 weakly similar to PIR|T10798|T10798 pherophorin-S - Volvox carteri,
partial (6%)
Length = 957
Score = 34.7 bits (78), Expect = 0.064
Identities = 31/146 (21%), Positives = 65/146 (44%), Gaps = 19/146 (13%)
Frame = -3
Query: 255 QSDSIMHLTKSKINELVAILKDVES-------AQLRVAWLRSALD-----EIAENVEVIN 302
+++ + + K K + ++K+VE ++ RV L + EI E + I
Sbjct: 916 EAEGLKEVLKDKEGRVEELVKEVEGLKKVKAESEARVKDLEKRIGVLEMKEIEERNKRIR 737
Query: 303 QHQEVDAAKANTDREMESLREQLESEMESLAEKEQEVAD-------IKKRIPEISDRLKE 355
+E+ DRE++ R ++E + AEK+ E D +K + E ++ K
Sbjct: 736 VEEELRDTIGEKDREIDGFRNKVEELEKVGAEKKDEAGDWLNEKLSYEKALRESEEKAKG 557
Query: 356 LELKSAELEQSMLSSKSEVENLHRKS 381
E + +L + + S +++L+ K+
Sbjct: 556 FESQIVQLLEEVGESGKMIKSLNEKA 479
>TC93536 weakly similar to GP|2961371|emb|CAA18118.1 hypothetical protein
(fragment) {Arabidopsis thaliana}, partial (58%)
Length = 815
Score = 34.7 bits (78), Expect = 0.064
Identities = 29/129 (22%), Positives = 61/129 (46%), Gaps = 4/129 (3%)
Frame = +2
Query: 249 FVVQELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQH---- 304
+V ++S + M + ++ + +KD+E A L +A E+ ++ QH
Sbjct: 191 YVQISMESYTRMSGLEDQVVNMENQIKDLE------ANLSAAYSELDNKESLVKQHAKVA 352
Query: 305 QEVDAAKANTDREMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELE 364
+E + D E+ SLR QLES S ++ +A + + E +++ ++ +S +
Sbjct: 353 EEAVSGWEKADAEVVSLRHQLESITLSKLSCDERIAHLDGALKECMKQIRTVKEESEQKI 532
Query: 365 QSMLSSKSE 373
Q ++ KS+
Sbjct: 533 QEVILMKSQ 559
>TC85347 similar to GP|22136328|gb|AAM91242.1 unknown protein {Arabidopsis
thaliana}, partial (9%)
Length = 698
Score = 34.7 bits (78), Expect = 0.064
Identities = 12/39 (30%), Positives = 25/39 (63%)
Frame = +2
Query: 11 NPNCPHASNPYHQCTQACSKKTSHRTKPHAKNTSSSGAA 49
+P+C +ASNP+H+CT C + + ++ + +SG++
Sbjct: 392 HPDCRNASNPFHECTDYCFRVIAQAKLTQSEVSQASGSS 508
>TC88259 similar to GP|6729037|gb|AAF27033.1| unknown protein {Arabidopsis
thaliana}, partial (35%)
Length = 1757
Score = 33.9 bits (76), Expect = 0.11
Identities = 37/135 (27%), Positives = 57/135 (41%), Gaps = 16/135 (11%)
Frame = +3
Query: 264 KSKINELVAILKDVESAQLRVAWLRSALDEI------AENVEVINQHQEVDAAK------ 311
K+ NEL A L ++ + + V +E AEN EV + Q++DAA+
Sbjct: 429 KTLTNELAAALLEISAKEDMVKQHSKVAEEAISGWEKAEN-EVSSLKQQLDAARQKNSGL 605
Query: 312 ----ANTDREMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSM 367
++ D ++ QL E +K E + + R ELE K AELE +
Sbjct: 606 EDRVSHLDGALKECMRQLRQAREVQEQKIHEA--VANNSHDSGSRRFELERKVAELEAQL 779
Query: 368 LSSKSEVENLHRKSL 382
+SK+E R L
Sbjct: 780 QTSKAEAAASIRSDL 824
>TC93234 weakly similar to GP|21751020|dbj|BAC03887. unnamed protein product
{Homo sapiens}, partial (11%)
Length = 591
Score = 32.7 bits (73), Expect = 0.24
Identities = 34/118 (28%), Positives = 48/118 (39%), Gaps = 7/118 (5%)
Frame = +2
Query: 5 SNGTASNPNCPHA-SNPYHQCTQACSKKTSHRTKPHAKNTSSSGAA------PPPSNFDR 57
S GTA P A S+P T S ++S + + S+S +A PPP+ F
Sbjct: 248 STGTAP----PFAVSSPASSPTTPSSTRSSPGSTSTPRTPSTSFSARPPPSTPPPTPFRS 415
Query: 58 RKKPGSRPEPPVVAGAVPAAKVEAIQKSNASSPISNHSEIKKVESRNDDHNHSVPVSG 115
R GS P + + PA AI S S+P S S +S+ +SG
Sbjct: 416 RPTKGSSPSSTPSSSSPPAPTSPAISPSRPSAPTRRPSPPGTACSPRSRPPNSIVISG 589
>TC85063 similar to PIR|T47959|T47959 hypothetical protein F15G16.60 -
Arabidopsis thaliana, partial (5%)
Length = 742
Score = 32.7 bits (73), Expect = 0.24
Identities = 18/66 (27%), Positives = 32/66 (48%), Gaps = 1/66 (1%)
Frame = +3
Query: 39 HAKNTSSSGAAP-PPSNFDRRKKPGSRPEPPVVAGAVPAAKVEAIQKSNASSPISNHSEI 97
H N G+ P PP+ F + P + +P + +PAA + + S P++N E
Sbjct: 225 HCYNNKIRGSMPVPPATFLNSRFPHTSNDPMLYRNEIPAAVSQHVHNSRTGVPVANSRE- 401
Query: 98 KKVESR 103
K++ +R
Sbjct: 402 KQLHTR 419
>TC79642 similar to PIR|T07397|T07397 kinesin heavy chain-like protein
(clone PKCBP) - potato, partial (42%)
Length = 2321
Score = 32.7 bits (73), Expect = 0.24
Identities = 31/139 (22%), Positives = 61/139 (43%), Gaps = 8/139 (5%)
Frame = +2
Query: 250 VVQELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEVDA 309
V + Q + + K+ L A+ D E LRS ++ + ++ I + D
Sbjct: 308 VKMQEQLEGLKESLKANKQNLQAVTSDCER-------LRSLCNKKDQALQAIESTSKKDL 466
Query: 310 AKANTDREMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAE------- 362
+ N ++ ++ L+ +L+ L E+ + ++ + +L LE +++E
Sbjct: 467 VETN-NQVLQKLKYELKYCKGELDSAEETIKTLRSEKAILEQKLSVLEKRNSEESSSLLR 643
Query: 363 -LEQSMLSSKSEVENLHRK 380
LEQ + KSEV +L RK
Sbjct: 644 KLEQERKAVKSEVYDLERK 700
>AW697261 similar to GP|14596027|gb| Unknown protein {Arabidopsis thaliana},
partial (3%)
Length = 1372
Score = 32.7 bits (73), Expect = 0.24
Identities = 14/33 (42%), Positives = 17/33 (51%)
Frame = -1
Query: 36 TKPHAKNTSSSGAAPPPSNFDRRKKPGSRPEPP 68
T PH+ SS PPP R +P +RP PP
Sbjct: 121 TPPHSWKRSSPSYPPPPGISGSRTRPAARPPPP 23
>TC78089 similar to GP|4522009|gb|AAD21782.1| unknown protein {Arabidopsis
thaliana}, partial (85%)
Length = 2853
Score = 32.3 bits (72), Expect = 0.32
Identities = 31/150 (20%), Positives = 71/150 (46%)
Frame = +1
Query: 204 PVGKYHVKESFASILESILDKYGDIGASCHLESLVLRSYYIECVCFVVQELQSDSIMHLT 263
P+ H+ + S+L+ + + DIG S + +L +E C V + D ++
Sbjct: 694 PITSIHISTTCTSLLKELEQIWNDIGESEKDKDRMLME--LERECLDVYRRKVDEAANI- 864
Query: 264 KSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEVDAAKANTDREMESLRE 323
K++ ++ +A + + +A L +AL E ++ +++ A+ ++ S+
Sbjct: 865 KARFHQSLA------AKEAEIATLIAALGE-----HDMSSPIKMEKRSASLKEKLASITP 1011
Query: 324 QLESEMESLAEKEQEVADIKKRIPEISDRL 353
+E + E+ +++ADIK +I +IS +
Sbjct: 1012LVEELKKKKEERLKQLADIKTQIEKISGEI 1101
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.308 0.126 0.345
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,084,411
Number of Sequences: 36976
Number of extensions: 164174
Number of successful extensions: 1302
Number of sequences better than 10.0: 95
Number of HSP's better than 10.0 without gapping: 1218
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1284
length of query: 387
length of database: 9,014,727
effective HSP length: 98
effective length of query: 289
effective length of database: 5,391,079
effective search space: 1558021831
effective search space used: 1558021831
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0017.9


