

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
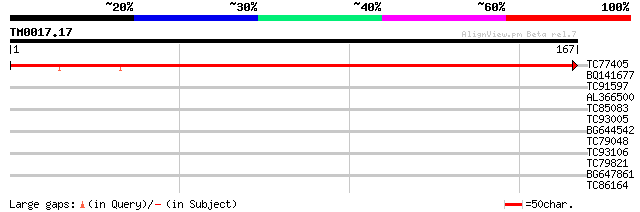
Query= TM0017.17
(167 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77405 similar to GP|21360368|gb|AAM47506.1 iron-stress related... 250 2e-67
BQ141677 28 1.3
TC91597 similar to GP|21323350|dbj|BAB97978. Hypothetical protei... 28 2.2
AL366500 27 2.9
TC85083 similar to GP|9758917|dbj|BAB09454.1 gene_id:MXI22.6~unk... 27 3.8
TC93005 27 5.0
BG644542 similar to PIR|D84681|D846 hypothetical protein At2g281... 27 5.0
TC79048 weakly similar to GP|15450639|gb|AAK96591.1 AT5g62520/K1... 27 5.0
TC93106 weakly similar to PIR|C96797|C96797 unknown protein [imp... 26 6.5
TC79821 similar to GP|6681331|gb|AAF23248.1| B' regulatory subun... 26 6.5
BG647861 similar to GP|6815055|dbj EST AU081527(C53732) correspo... 26 8.5
TC86164 weakly similar to GP|20514261|gb|AAM22959.1 tubulin fold... 26 8.5
>TC77405 similar to GP|21360368|gb|AAM47506.1 iron-stress related protein
{Citrus junos}, partial (13%)
Length = 944
Score = 250 bits (638), Expect = 2e-67
Identities = 129/172 (75%), Positives = 139/172 (80%), Gaps = 5/172 (2%)
Frame = +3
Query: 1 MLENLHPAPPSES---VKRYAPPNQRNRSTNRRK--SSDRLDRTNSIGNDLEKNQFASSR 55
MLEN PP VKRYAPPNQRNRS N R+ SSDRLDR+NS+G+D+EKNQ ASS+
Sbjct: 75 MLENPQAQPPESGPVVVKRYAPPNQRNRSINNRRKSSSDRLDRSNSVGSDIEKNQVASSK 254
Query: 56 NVQVQDRIDAVSSNLVNENPYSRFVALDGCSCSAASQLLNDRWTAAIQSYNNSKDSPEKP 115
N+QV D A SSN+ NEN YSRF+AL GCS SAASQLL+DRWTA IQSYNN KDS EKP
Sbjct: 255 NIQVTDHGHASSSNIFNENHYSRFIALQGCSSSAASQLLSDRWTAVIQSYNNPKDSSEKP 434
Query: 116 VMYSGGTSVWTHFRLPHQIMSPAASAPPSVSQLDFLGELNRQMQSVNPSFST 167
VMYSGGTSVWT FRLPHQIMSPAASA SVSQLDFLGEL RQM S NPSFST
Sbjct: 435 VMYSGGTSVWTQFRLPHQIMSPAASASSSVSQLDFLGELRRQMHSANPSFST 590
>BQ141677
Length = 910
Score = 28.5 bits (62), Expect = 1.3
Identities = 15/42 (35%), Positives = 23/42 (54%)
Frame = +3
Query: 104 SYNNSKDSPEKPVMYSGGTSVWTHFRLPHQIMSPAASAPPSV 145
+Y++ P KP++ +GGTS F +S A S PPS+
Sbjct: 57 TYDSDSSQPLKPLLLAGGTSTLLLF------ISAAESPPPSL 164
>TC91597 similar to GP|21323350|dbj|BAB97978. Hypothetical protein
{Corynebacterium glutamicum ATCC 13032}, partial (5%)
Length = 718
Score = 27.7 bits (60), Expect = 2.2
Identities = 20/71 (28%), Positives = 37/71 (51%), Gaps = 2/71 (2%)
Frame = +3
Query: 10 PSESVKRYAPPNQRNRSTN--RRKSSDRLDRTNSIGNDLEKNQFASSRNVQVQDRIDAVS 67
P +S +R ++RN S N R + +D T ++ +L +++ + SRN+ RID +
Sbjct: 267 PPQSWRRNLLRSRRNLSRNLPRNRRTDPSRSTRNLLRNLSRSRRSLSRNLLRSRRIDLLR 446
Query: 68 SNLVNENPYSR 78
S N + +R
Sbjct: 447 SRRRNPSRSTR 479
>AL366500
Length = 520
Score = 27.3 bits (59), Expect = 2.9
Identities = 11/29 (37%), Positives = 16/29 (54%)
Frame = -1
Query: 49 NQFASSRNVQVQDRIDAVSSNLVNENPYS 77
N+F SRN+ +Q N++N N YS
Sbjct: 436 NRFH*SRNIYIQSFFQKKEKNIINHNSYS 350
>TC85083 similar to GP|9758917|dbj|BAB09454.1 gene_id:MXI22.6~unknown
protein {Arabidopsis thaliana}, partial (8%)
Length = 752
Score = 26.9 bits (58), Expect = 3.8
Identities = 17/38 (44%), Positives = 20/38 (51%)
Frame = +3
Query: 11 SESVKRYAPPNQRNRSTNRRKSSDRLDRTNSIGNDLEK 48
SES A +R RS +RR S DR NSIG +K
Sbjct: 459 SESESTPASSLRRGRSVSRRSSGVGDDRRNSIGGGGKK 572
>TC93005
Length = 702
Score = 26.6 bits (57), Expect = 5.0
Identities = 10/30 (33%), Positives = 17/30 (56%)
Frame = -3
Query: 126 THFRLPHQIMSPAASAPPSVSQLDFLGELN 155
TH ++PH ++ A P +S +DF+ N
Sbjct: 559 THLQIPHLLLFNAFRILPLLSSIDFISNTN 470
>BG644542 similar to PIR|D84681|D846 hypothetical protein At2g28150
[imported] - Arabidopsis thaliana, partial (5%)
Length = 781
Score = 26.6 bits (57), Expect = 5.0
Identities = 15/62 (24%), Positives = 29/62 (46%), Gaps = 4/62 (6%)
Frame = -3
Query: 18 APPNQRNRSTNRRKSSDRLDRTNSIG----NDLEKNQFASSRNVQVQDRIDAVSSNLVNE 73
+PP++ +RS S DRTN G + F + VQ+++R+ + + ++
Sbjct: 422 SPPSEFSRSPISDGSRSSRDRTNDSGIRQKQSTRLDSFRDEKVVQIEERLASGARVIIQS 243
Query: 74 NP 75
P
Sbjct: 242 KP 237
>TC79048 weakly similar to GP|15450639|gb|AAK96591.1 AT5g62520/K19B1_13
{Arabidopsis thaliana}, partial (19%)
Length = 1709
Score = 26.6 bits (57), Expect = 5.0
Identities = 14/38 (36%), Positives = 20/38 (51%)
Frame = -3
Query: 5 LHPAPPSESVKRYAPPNQRNRSTNRRKSSDRLDRTNSI 42
L+P PPS S + Y ++ R + RKS L R N +
Sbjct: 573 LYPHPPSSSEQVYWNRGRQMRHSRNRKSLIHLRRRNHV 460
>TC93106 weakly similar to PIR|C96797|C96797 unknown protein [imported] -
Arabidopsis thaliana, partial (21%)
Length = 792
Score = 26.2 bits (56), Expect = 6.5
Identities = 19/68 (27%), Positives = 34/68 (49%), Gaps = 1/68 (1%)
Frame = +3
Query: 9 PPSESVKRYAP-PNQRNRSTNRRKSSDRLDRTNSIGNDLEKNQFASSRNVQVQDRIDAVS 67
P S+ V Y NQR R + + D R ++ N+L + + +R Q+Q+ + + S
Sbjct: 45 PESDPVWHYLNIQNQRIRGLLEQCTLDHEARMENLRNELHEKALSDARWKQIQEEL-SES 221
Query: 68 SNLVNENP 75
S++ N P
Sbjct: 222 SDINNSYP 245
>TC79821 similar to GP|6681331|gb|AAF23248.1| B' regulatory subunit of PP2A
(AtB'beta) {Arabidopsis thaliana}, partial (36%)
Length = 860
Score = 26.2 bits (56), Expect = 6.5
Identities = 12/30 (40%), Positives = 17/30 (56%)
Frame = -3
Query: 73 ENPYSRFVALDGCSCSAASQLLNDRWTAAI 102
ENP + + L+G SC ASQ + RW +
Sbjct: 198 ENPNPKLMNLEGESCDVASQ-VKKRWIGVV 112
>BG647861 similar to GP|6815055|dbj EST AU081527(C53732) corresponds to a
region of the predicted gene.~hypothetical protein,
partial (72%)
Length = 733
Score = 25.8 bits (55), Expect = 8.5
Identities = 13/36 (36%), Positives = 19/36 (52%)
Frame = +2
Query: 84 GCSCSAASQLLNDRWTAAIQSYNNSKDSPEKPVMYS 119
GCSCS+ + N + I S+ KDS K ++S
Sbjct: 305 GCSCSSTACFFNLPRSTVITSFRR*KDSEIKNFIHS 412
>TC86164 weakly similar to GP|20514261|gb|AAM22959.1 tubulin folding
cofactor C {Arabidopsis thaliana}, partial (8%)
Length = 1369
Score = 25.8 bits (55), Expect = 8.5
Identities = 11/27 (40%), Positives = 16/27 (58%)
Frame = +2
Query: 7 PAPPSESVKRYAPPNQRNRSTNRRKSS 33
P PP +V PP R RS+ RR+++
Sbjct: 575 PCPPHTAVPSSPPPPPRMRSSPRRRTT 655
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.311 0.125 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,233,385
Number of Sequences: 36976
Number of extensions: 67601
Number of successful extensions: 310
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 306
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 309
length of query: 167
length of database: 9,014,727
effective HSP length: 89
effective length of query: 78
effective length of database: 5,723,863
effective search space: 446461314
effective search space used: 446461314
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0017.17


