

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
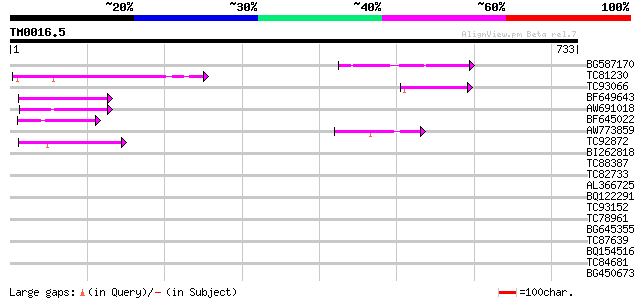
Query= TM0016.5
(733 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 122 6e-28
TC81230 100 2e-21
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 78 1e-14
BF649643 similar to PIR|H86486|H86 protein Ty1/copia-element pol... 72 1e-12
AW691018 weakly similar to GP|2462935|em open reading frame 1 {B... 63 4e-10
BF645022 weakly similar to GP|8777581|dbj retroelement pol polyp... 53 5e-07
AW773859 44 2e-04
TC92872 42 6e-04
BI262818 weakly similar to GP|6642775|gb| gag-pol polyprotein {V... 42 0.001
TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finge... 40 0.002
TC82733 similar to GP|10177404|dbj|BAB10535. gene_id:K24M7.12~pi... 35 0.079
AL366725 35 0.10
BQ122291 35 0.10
TC93152 34 0.23
TC78961 similar to GP|18252179|gb|AAL61922.1 unknown protein {Ar... 34 0.23
BG645355 similar to PIR|G96590|G965 hypothetical protein T24C10.... 34 0.23
TC87639 GP|9663153|emb|CAC01132.1 transport-secretion protein 2.... 33 0.30
BQ154516 33 0.51
TC84681 similar to GP|1370182|emb|CAA98168.1 RAB7A {Lotus japoni... 31 1.5
BG450673 homologue to GP|12055499|emb NADH dehydrogenase subunit... 31 1.5
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 122 bits (305), Expect = 6e-28
Identities = 72/175 (41%), Positives = 95/175 (54%)
Frame = -3
Query: 426 SGKLIARGPKVGRLFPLCFSMSPCSSLPFVSCHSATVVNFELWHKRLGHPNSNVLYELLQ 485
S +LI G G L+ L + P S+ S+++ LWH RLGHP+ L +L
Sbjct: 704 SSQLIGEGVTKGDLYML-EKLDPVSNYKCSFTSSSSLNKDALWHARLGHPHGRALNLMLP 528
Query: 486 SGVLGNKETPSLSTIKFDCTYCKLGKSKILPFPNHQSNISQTFDMIHSDLWGITPVISHA 545
V NK +C C LGK FP + FD+I++DLW P +S
Sbjct: 527 GVVFENK----------NCEACILGKHCKNVFPRTSTVYENCFDLIYTDLW-TAPSLSRD 381
Query: 546 NYKYFVTFIDDFSRFTWVYFLRSKDEVFSAFKFFHAYVETQFSSKIKILRSDNGG 600
N+KYFVTFID+ S++TW+ + SKD V AFK F AYV + +KIKILRSDNGG
Sbjct: 380 NHKYFVTFIDEKSKYTWLTLIPSKDRVIDAFKNFQAYVTNHYHAKIKILRSDNGG 216
>TC81230
Length = 958
Score = 100 bits (250), Expect = 2e-21
Identities = 71/264 (26%), Positives = 120/264 (44%), Gaps = 10/264 (3%)
Frame = +1
Query: 4 EKYEV---LLVRLNGKNYSAWAFEFQIFVKGKSLWGHVDGSTSALDKEKQETEYA----- 55
EK EV + + LNG NY+ WA F+KG+ LW +V G K K +T A
Sbjct: 202 EKLEVTPPISIILNGSNYNHWAESMCGFLKGRRLWRYVTGDKKCPTKGKDDTADAFADKL 381
Query: 56 -DWEVKDNQVLAWIIRFVDPNIVLNLRPFSTAAKMWAYLKKIYNQNNTARRFQLEHDIA- 113
+W+ K++Q++ W P+I + F A ++W +LK+ Y ++ + ++QL D++
Sbjct: 382 EEWDSKNHQIITWFRNTSIPSIHMQFGRFENAKEVWDHLKQRYTISDLSHQYQLLKDLSN 561
Query: 114 IFQQEILSISEFYSQFMNLWADYTDIVYGSVSTEGLTSVQTVHETTK*DQFLMKLRSDFE 173
+ QQ + EF +Q +W T + + +T + QFLM L ++E
Sbjct: 562 LKQQSGQPVYEFLAQMEVIWNQLTSCEPSLKDATDMKTYETHRNRVRLIQFLMALTDEYE 741
Query: 174 GIRTNLMNRATVPS*DACLNELLREERRLLTLATMEQHKSASLPVAYVVQEKPRGRDLSA 233
+R + +++ +P+ + L L EE RL Q +A+ V +
Sbjct: 742 PVRASSLHQNPLPTLENALPCLKSEETRL-------QLVPPKADLAFAVTNN------AT 882
Query: 234 VQCFCCKGFGHYASNCPRKSCNYC 257
C C+ GH S+CP C C
Sbjct: 883 KPCRHCQKSGHSFSDCPTIECRNC 954
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 77.8 bits (190), Expect = 1e-14
Identities = 41/97 (42%), Positives = 51/97 (52%), Gaps = 4/97 (4%)
Frame = +1
Query: 506 YCK----LGKSKILPFPNHQSNISQTFDMIHSDLWGITPVISHANYKYFVTFIDDFSRFT 561
+CK G K + F D IHSDLWG + V S+ +Y +T IDDF R
Sbjct: 19 FCKHLLFFGNRKKVSFSTATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKV 198
Query: 562 WVYFLRSKDEVFSAFKFFHAYVETQFSSKIKILRSDN 598
WVYFLR K+E F FK + VETQ +K L +DN
Sbjct: 199 WVYFLRYKNETFPTFKKWRILVETQTGKNVKKLITDN 309
>BF649643 similar to PIR|H86486|H86 protein Ty1/copia-element polyprotein
[imported] - Arabidopsis thaliana, partial (2%)
Length = 608
Score = 71.6 bits (174), Expect = 1e-12
Identities = 33/122 (27%), Positives = 63/122 (51%)
Frame = +1
Query: 12 RLNGKNYSAWAFEFQIFVKGKSLWGHVDGSTSALDKEKQETEYADWEVKDNQVLAWIIRF 71
+LNG NYS+W+ + K+ G ++GS + Q EYA W ++ +L+W+
Sbjct: 172 KLNGTNYSSWSRSMVHALTAKNKVGFINGSIKTPSEVDQPAEYALWNRCNSMILSWLTHS 351
Query: 72 VDPNIVLNLRPFSTAAKMWAYLKKIYNQNNTARRFQLEHDIAIFQQEILSISEFYSQFMN 131
V+P++ + TA ++W K ++Q N +Q++ +A Q +S S ++++
Sbjct: 352 VEPDLAKGVIHAKTACQVWEDFKDQFSQKNIPAIYQIQKSLASLSQGTVSASTYFTKIKG 531
Query: 132 LW 133
LW
Sbjct: 532 LW 537
>AW691018 weakly similar to GP|2462935|em open reading frame 1 {Brassica
oleracea}, partial (12%)
Length = 639
Score = 63.2 bits (152), Expect = 4e-10
Identities = 33/120 (27%), Positives = 66/120 (54%)
Frame = +2
Query: 13 LNGKNYSAWAFEFQIFVKGKSLWGHVDGSTSALDKEKQETEYADWEVKDNQVLAWIIRFV 72
LNG NY W+ + + K+ G +DG+ A + + W+ ++ VL WI+ +
Sbjct: 209 LNGGNYGEWSRSMLLSLSAKNKLGLIDGTVKAPSADDPKLPL--WKRCNDLVLTWILHSI 382
Query: 73 DPNIVLNLRPFSTAAKMWAYLKKIYNQNNTARRFQLEHDIAIFQQEILSISEFYSQFMNL 132
+P+I ++ TAA +W+ L ++Q + +R +Q+ +I+ +Q L IS++Y++ +L
Sbjct: 383 EPDIARSVIFSDTAAAVWSDLHDRFSQGDESRIYQIRQEISECRQGSLLISDYYTKLKSL 562
>BF645022 weakly similar to GP|8777581|dbj retroelement pol polyprotein-like
{Arabidopsis thaliana}, partial (8%)
Length = 685
Score = 52.8 bits (125), Expect = 5e-07
Identities = 26/107 (24%), Positives = 53/107 (49%)
Frame = -2
Query: 11 VRLNGKNYSAWAFEFQIFVKGKSLWGHVDGSTSALDKEKQETEYADWEVKDNQVLAWIIR 70
++L G NY W+ + + K +G V+G + K + DW+ + ++AW++
Sbjct: 330 IQLRGLNYDEWSRAIRTSFQAKRKYGFVEGK---IPKPTTPEKLEDWKAVQSMLIAWLLN 160
Query: 71 FVDPNIVLNLRPFSTAAKMWAYLKKIYNQNNTARRFQLEHDIAIFQQ 117
++P++ L + A +W +LK+ + N AR QL+ + +Q
Sbjct: 159 TIEPSLRSTLSYYDDAESLWTHLKQRFCVVNGARICQLKASLGECKQ 19
>AW773859
Length = 538
Score = 43.9 bits (102), Expect = 2e-04
Identities = 31/122 (25%), Positives = 52/122 (42%), Gaps = 4/122 (3%)
Frame = -3
Query: 420 LVQDQHSGKLIARGPKVGRLFPLCFSMSPCSSLPFVSCHSATVVNF----ELWHKRLGHP 475
L+Q++ S K+I G L+ L +P +++ F LWH RLGH
Sbjct: 383 LIQEKRSLKMIGSGELFEGLYFLTTQDTPSATIASSQVQPQPQPTFLPQEALWHFRLGHL 204
Query: 476 NSNVLYELLQSGVLGNKETPSLSTIKFDCTYCKLGKSKILPFPNHQSNISQTFDMIHSDL 535
++ L L + + S+ C C + K LPF + S+ +++ H D+
Sbjct: 203 SNRKLLSLHSNFPFITIDQNSV------CDICHYSRHKKLPFQLSTNRASKCYELFHFDI 42
Query: 536 WG 537
WG
Sbjct: 41 WG 36
>TC92872
Length = 923
Score = 42.4 bits (98), Expect = 6e-04
Identities = 27/146 (18%), Positives = 63/146 (42%), Gaps = 7/146 (4%)
Frame = +1
Query: 12 RLNGKNYSAWAFEFQIFVKGKSLWGHVDGSTSALDK-------EKQETEYADWEVKDNQV 64
+L+ KN+ AW + + G + GH+DG+T + +Y W D +
Sbjct: 106 KLSRKNFRAWKRQVTTLLAGIEVMGHIDGTTPIQTETIINNGVSAPNPDYTKWFTLDQLI 285
Query: 65 LAWIIRFVDPNIVLNLRPFSTAAKMWAYLKKIYNQNNTARRFQLEHDIAIFQQEILSISE 124
+ ++ + ++ + TA +W ++ +N + + + + I + SI++
Sbjct: 286 INLLLSSMTEADSISFASYETARTLWVAIESQFNNTSRSHVMSVTNQIQRCTKGDKSITD 465
Query: 125 FYSQFMNLWADYTDIVYGSVSTEGLT 150
+ +L AD ++ S+S + +T
Sbjct: 466 YLFSVKSL-ADELAVIDKSLSDDDIT 540
>BI262818 weakly similar to GP|6642775|gb| gag-pol polyprotein {Vitis
vinifera}, partial (24%)
Length = 615
Score = 41.6 bits (96), Expect = 0.001
Identities = 25/123 (20%), Positives = 51/123 (41%), Gaps = 5/123 (4%)
Frame = +1
Query: 14 NGKNYSAWAFEFQIFVKGKSLWGHVDGSTSALDKEKQET-----EYADWEVKDNQVLAWI 68
+G+NY WA + +++ LW V+ L T + + + + ++ A +
Sbjct: 187 DGENYHIWAARIEAYLEANDLWEAVEEDYEVLPLSDNPTMAQIKNHKERKTRKSKARATL 366
Query: 69 IRFVDPNIVLNLRPFSTAAKMWAYLKKIYNQNNTARRFQLEHDIAIFQQEILSISEFYSQ 128
V I + +A ++W +LK Y + R Q + I F+ + + SE +
Sbjct: 367 FAAVSEEIFTRIMTIKSAFEIWNFLKTEYEGDERIRGMQALNLIREFEMQKMKESETIKE 546
Query: 129 FMN 131
+ N
Sbjct: 547 YAN 555
>TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finger protein
{Oryza sativa (japonica cultivar-group)}, partial (96%)
Length = 1286
Score = 40.4 bits (93), Expect = 0.002
Identities = 19/40 (47%), Positives = 25/40 (62%), Gaps = 3/40 (7%)
Frame = +1
Query: 236 CFCCKGFGHYASNCPRKS-CNYCKKDGHVIKEC--PIRPP 272
C+ CK GH AS+CP + C+ C K GH +EC P +PP
Sbjct: 538 CWNCKEPGHMASSCPNEGICHTCGKAGHRARECTVPQKPP 657
Score = 39.3 bits (90), Expect = 0.005
Identities = 22/51 (43%), Positives = 28/51 (54%), Gaps = 1/51 (1%)
Frame = +1
Query: 222 VQEKPRGRDLSAVQCFCCKGFGHYASNCPR-KSCNYCKKDGHVIKECPIRP 271
V +KP G DL C C GH A C K+CN C+K GH+ ++CP P
Sbjct: 640 VPQKPPG-DLRL--CNNCYKQGHIAVECTNEKACNNCRKTGHLARDCPNDP 783
Score = 37.7 bits (86), Expect = 0.016
Identities = 15/34 (44%), Positives = 19/34 (55%), Gaps = 1/34 (2%)
Frame = +1
Query: 236 CFCCKGFGHYASNCPRKS-CNYCKKDGHVIKECP 268
C C+ GH A +CP CN C GHV ++CP
Sbjct: 730 CNNCRKTGHLARDCPNDPICNLCNISGHVARQCP 831
Score = 34.7 bits (78), Expect = 0.13
Identities = 16/34 (47%), Positives = 18/34 (52%), Gaps = 1/34 (2%)
Frame = +1
Query: 236 CFCCKGFGHYASNCPRKS-CNYCKKDGHVIKECP 268
C C GH AS C KS C CK+ GH+ CP
Sbjct: 481 CHNCSLPGHIASECSTKSLCWNCKEPGHMASSCP 582
Score = 34.7 bits (78), Expect = 0.13
Identities = 14/33 (42%), Positives = 17/33 (51%), Gaps = 1/33 (3%)
Frame = +1
Query: 236 CFCCKGFGHYASNCPRKS-CNYCKKDGHVIKEC 267
C CK GHY CP + C+ C GH+ EC
Sbjct: 424 CKNCKRPGHYVRECPNVAVCHNCSLPGHIASEC 522
>TC82733 similar to GP|10177404|dbj|BAB10535.
gene_id:K24M7.12~pir||S42136~similar to unknown protein
{Arabidopsis thaliana}, partial (57%)
Length = 710
Score = 35.4 bits (80), Expect = 0.079
Identities = 15/40 (37%), Positives = 20/40 (49%), Gaps = 7/40 (17%)
Frame = +3
Query: 236 CFCCKGFGHYASNCPRKS-------CNYCKKDGHVIKECP 268
CF CKG H A C +K+ C C++ GH + CP
Sbjct: 282 CFICKGLDHIAKFCTQKAEWEKNKICLRCRRRGHRAQNCP 401
Score = 31.6 bits (70), Expect = 1.1
Identities = 14/39 (35%), Positives = 17/39 (42%), Gaps = 9/39 (23%)
Frame = +3
Query: 239 CKGFGHYASNCPRK---------SCNYCKKDGHVIKECP 268
C GH +NCP C CK+ GH+ K CP
Sbjct: 441 CGDNGHSLANCPHPLQEGGTMFAQCFVCKEQGHLSKNCP 557
Score = 31.6 bits (70), Expect = 1.1
Identities = 12/27 (44%), Positives = 15/27 (55%)
Frame = +3
Query: 235 QCFCCKGFGHYASNCPRKSCNYCKKDG 261
QCF CK GH + NCP+ + K G
Sbjct: 510 QCFVCKEQGHLSKNCPKNAHGIYPKGG 590
>AL366725
Length = 485
Score = 35.0 bits (79), Expect = 0.10
Identities = 17/50 (34%), Positives = 27/50 (54%), Gaps = 3/50 (6%)
Frame = +2
Query: 221 VVQEKPRGRDLSA-VQCFCCKGFGHYASNCPR--KSCNYCKKDGHVIKEC 267
V +P+ +D A + CF GH ++ CP+ K C C K GH++ +C
Sbjct: 260 VDDRRPKKKDAPAEIVCFNYGEKGHKSNVCPKEIKKCVRCDKKGHIVADC 409
>BQ122291
Length = 487
Score = 35.0 bits (79), Expect = 0.10
Identities = 30/149 (20%), Positives = 62/149 (41%), Gaps = 1/149 (0%)
Frame = -2
Query: 54 YADWEVKDNQVLAWIIRFVDPNIVLNLRPFSTAAKMWAYLKKIYNQNNTARRFQLEHDIA 113
Y +WE KD+ + WI+ + P+++ + ++W + R QL ++
Sbjct: 450 YTEWEDKDSLLCTWILSRISPSLLSRFVLLRHSWQVWDEIHSYCFTQMKTRSRQLRSELR 271
Query: 114 IFQQEILSISEFYSQFMNLWADYTDIVYGSVSTEGLTSV-QTVHETTK*DQFLMKLRSDF 172
+ ++SEF ++ + +E L S+ V + L L +F
Sbjct: 270 SITKGSRTVSEFIARIRAI-------------SESLASIGDPVSHRDLIEVVLEALPEEF 130
Query: 173 EGIRTNLMNRATVPS*DACLNELLREERR 201
+ I ++ ++ V S D ++LL +E R
Sbjct: 129 DPIVASVNAKSEVVSLDELESQLLTQESR 43
>TC93152
Length = 647
Score = 33.9 bits (76), Expect = 0.23
Identities = 23/92 (25%), Positives = 42/92 (45%), Gaps = 1/92 (1%)
Frame = +3
Query: 13 LNGKNYSAWAFEFQIFVKGKSLWGHVDG-STSALDKEKQETEYADWEVKDNQVLAWIIRF 71
L NY W+ + ++ G+ LWG V S S + ++ETE W ++ + L I
Sbjct: 150 LTKDNYDYWSCLVRNYLLGQDLWGFVTSISESTGPRSRRETEV--WNRRNGKALHIIQLA 323
Query: 72 VDPNIVLNLRPFSTAAKMWAYLKKIYNQNNTA 103
P + ++ +A + W L Y+ + +A
Sbjct: 324 CGPENINLIKDLQSAREAWNELSTHYSSDLSA 419
>TC78961 similar to GP|18252179|gb|AAL61922.1 unknown protein {Arabidopsis
thaliana}, partial (71%)
Length = 974
Score = 33.9 bits (76), Expect = 0.23
Identities = 17/51 (33%), Positives = 24/51 (46%), Gaps = 4/51 (7%)
Frame = +2
Query: 227 RGRDLSAVQCFCCKGFGHYASNCP----RKSCNYCKKDGHVIKECPIRPPK 273
RG + +CF C GH+A +C + C C + GH+ K C P K
Sbjct: 332 RGPPPGSGRCFNCGIDGHWARDCKAGDWKNKCYRCGERGHIEKNCKNSPKK 484
>BG645355 similar to PIR|G96590|G965 hypothetical protein T24C10.5 [imported]
- Arabidopsis thaliana, partial (5%)
Length = 627
Score = 33.9 bits (76), Expect = 0.23
Identities = 16/49 (32%), Positives = 21/49 (42%), Gaps = 15/49 (30%)
Frame = -1
Query: 235 QCFCCKGFGHYASNCPRKS---------------CNYCKKDGHVIKECP 268
+C+ C+ GH+ASNCP S C C + GH CP
Sbjct: 504 KCYKCQQPGHWASNCPSMSAANRVSGGSGGASGNCYKCNQPGHWANNCP 358
>TC87639 GP|9663153|emb|CAC01132.1 transport-secretion protein 2.2 (TTS-2.2)
{Homo sapiens}, partial (2%)
Length = 1522
Score = 33.5 bits (75), Expect = 0.30
Identities = 61/269 (22%), Positives = 100/269 (36%), Gaps = 25/269 (9%)
Frame = -1
Query: 30 KGKSLWGHVDGSTSALDKEKQETEYADWEVKDNQVLAWIIRFVDPNIVLNLRPFSTAA-- 87
K S W H +D++ ET+ W + + +DP + + F A
Sbjct: 964 KEYSAWAHQMRGKLKIDRDFFETDEQRWYL--------VYSCLDPGVQQVVETFFAAGGP 809
Query: 88 -------KMWAYLKKIYNQNNTARRFQLEHDIAIFQQEILSISEFYSQFMNLWAD----- 135
AYL + Y N R + ++ Q++ ++ F +F + AD
Sbjct: 808 GGNQDPQDFMAYLDRTYLDPNIQSRAVAQLQ-SLRQKDTERLATFLPRFEKVLADAGGYS 632
Query: 136 YTDIVYGSVSTEGLTS-----VQTVHETTK*DQFLMKLRSDFEGIRTNLMNRATVPS*DA 190
+ D+V S+ L + TV T Q+L K++ I + T P+ A
Sbjct: 631 WPDVVQISLLETALVPRLKELLITVELPTVYSQWLSKVQD----IAWKMERMKTPPTRWA 464
Query: 191 CLNELL----REERRLLTLATMEQHK--SASLPVAYVVQEKPRGRDLSAVQCFCCKGFGH 244
L R+ ++T A +Q + +S V+ P RD+ +C+ C GH
Sbjct: 463 PATRLPVSKDRDGDMMMTGAIHKQRRRRGSSSSVSSAEGAPPPRRDMR--ECYSCHERGH 290
Query: 245 YASNCPRKSCNYCKKDGHVIKECPIRPPK 273
A NC S KK G K + P K
Sbjct: 289 IARNCTNTSAAKKKKKGP--KVAKVEPQK 209
>BQ154516
Length = 506
Score = 32.7 bits (73), Expect = 0.51
Identities = 18/44 (40%), Positives = 27/44 (60%), Gaps = 4/44 (9%)
Frame = +1
Query: 445 SMSPCSS--LPFVSCHSATVVN--FELWHKRLGHPNSNVLYELL 484
S+S C++ + V C+S++ N F WH RLGH N NV+ +L
Sbjct: 325 SLSYCNNATISSVVCNSSSSSNDSFNKWHCRLGHANPNVVKSIL 456
Score = 30.4 bits (67), Expect = 2.5
Identities = 15/41 (36%), Positives = 22/41 (53%)
Frame = +3
Query: 331 SPNSSPWYFDYGASNHMANNAEALTNITQNFGNLKIQVANG 371
S + +PWY D GAS+H+ N L T G ++ + NG
Sbjct: 108 SYSEAPWYPDSGASHHLTFNPHNLAYRTPYNGQEQVLMDNG 230
>TC84681 similar to GP|1370182|emb|CAA98168.1 RAB7A {Lotus japonicus},
partial (63%)
Length = 856
Score = 31.2 bits (69), Expect = 1.5
Identities = 18/62 (29%), Positives = 28/62 (45%)
Frame = -1
Query: 456 SCHSATVVNFELWHKRLGHPNSNVLYELLQSGVLGNKETPSLSTIKFDCTYCKLGKSKIL 515
+C S T LW+ VL+ L+ G LGN + S++ + C K+G +
Sbjct: 406 ACTSTTTTLLTLWNSLRNSLKIYVLFTLIGQGSLGNTQECSINIVILFCRCLKVGN---I 236
Query: 516 PF 517
PF
Sbjct: 235 PF 230
>BG450673 homologue to GP|12055499|emb NADH dehydrogenase subunit 2 {Sphagnum
fallax}, partial (2%)
Length = 678
Score = 31.2 bits (69), Expect = 1.5
Identities = 16/62 (25%), Positives = 28/62 (44%)
Frame = +3
Query: 29 VKGKSLWGHVDGSTSALDKEKQETEYADWEVKDNQVLAWIIRFVDPNIVLNLRPFSTAAK 88
V GK G+++G + + + W + V WII +D +++ N FST
Sbjct: 222 VSGKDKLGYINGDLPP--PPQTDPTFRRWGTDNAIVKGWIINSMDSSLISNFIRFSTTKL 395
Query: 89 MW 90
+W
Sbjct: 396 VW 401
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.331 0.141 0.457
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,443,886
Number of Sequences: 36976
Number of extensions: 434563
Number of successful extensions: 2892
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 2129
Number of HSP's successfully gapped in prelim test: 92
Number of HSP's that attempted gapping in prelim test: 687
Number of HSP's gapped (non-prelim): 2297
length of query: 733
length of database: 9,014,727
effective HSP length: 103
effective length of query: 630
effective length of database: 5,206,199
effective search space: 3279905370
effective search space used: 3279905370
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0016.5


