

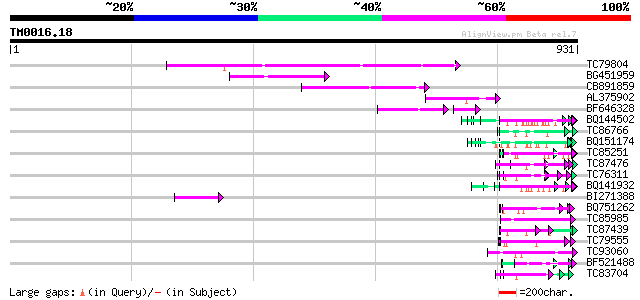
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0016.18
(931 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC79804 weakly similar to GP|6175165|gb|AAF04891.1| Mutator-like... 200 2e-51
BG451959 homologue to GP|6175165|gb| Mutator-like transposase {A... 99 6e-21
CB891859 similar to GP|6175165|gb| Mutator-like transposase {Ara... 90 3e-18
AL375902 similar to GP|9759134|dbj mutator-like transposase-like... 80 3e-15
BF646328 similar to PIR|F84533|F845 Mutator-like transposase [im... 57 5e-15
BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 67 4e-11
TC86766 weakly similar to PIR|T06291|T06291 extensin homolog T9E... 59 9e-09
BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xy... 58 1e-08
TC85251 homologue to PIR|JQ2128|JQ2128 metallothionein - soybean... 57 3e-08
TC87476 homologue to PIR|T11622|T11622 extensin class 1 precurso... 57 4e-08
TC76311 homologue to GP|3204132|emb|CAA07235.1 extensin {Cicer a... 57 4e-08
BQ141932 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 56 6e-08
BI271388 similar to GP|10177186|db mutator-like transposase-like... 56 6e-08
BQ751262 similar to GP|6715100|gb|A versicolorin B synthase {Asp... 56 7e-08
TC85985 similar to GP|5139695|dbj|BAA81686.1 expressed in cucumb... 55 1e-07
TC87439 weakly similar to GP|5139695|dbj|BAA81686.1 expressed in... 55 2e-07
TC79555 homologue to GP|10567859|gb|AAG18593.1 L8019.5 {Leishman... 54 2e-07
TC93060 weakly similar to PIR|S55970|S55970 ribosomal protein L6... 54 2e-07
BF521488 weakly similar to GP|18447433|gb RE28280p {Drosophila m... 54 3e-07
TC83704 similar to PIR|T05005|T05005 hypothetical protein T19P19... 54 4e-07
>TC79804 weakly similar to GP|6175165|gb|AAF04891.1| Mutator-like
transposase {Arabidopsis thaliana}, partial (57%)
Length = 2129
Score = 200 bits (509), Expect = 2e-51
Identities = 152/525 (28%), Positives = 229/525 (42%), Gaps = 42/525 (8%)
Frame = +1
Query: 258 DFDKDYKFRLGMEFASTEEFKKAILAHSVMNGKAVIYKKNDKNRVRFACKQEECRFIAYC 317
D +D+ F +G EF + F+ AI ++ + K+D R C E C +
Sbjct: 154 DCMEDHNFVVGQEFPDVKAFRNAIKEAAIAQHFELRIIKSDLIRYFAKCASEGCPWRIRA 333
Query: 318 ANIKGTSTYRIKTLNPDHTCGRTFKN--RAATAKWI------------------------ 351
+ ST+ I++L HTCGR N A+ WI
Sbjct: 334 VKLPNASTFTIRSLEGTHTCGRNALNGHHQASVDWIVSFIEERLRDNINYKPKDILHDIH 513
Query: 352 -----------AKNARKIAKEAVEGDALKQFTLITSYAAELMRVNHENTAAIILRYIPTT 400
A A++ A+ G + + F L+ S+ E+ + N + A + +
Sbjct: 514 KQYGITIPYKQAWRAKERGLAAIYGSSEEGFYLLPSFCEEIKKTNPGSVAKVFTTGADSR 693
Query: 401 LQPRFGCFYMCLEGCKSAFKLACRPFIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLAFA 460
Q F FY + G F C P + + G LK+KY L A D + FPLAFA
Sbjct: 694 FQRLFISFYASIHG----FVNGCLPIVALGGIQLKSKYLSTFLSATSFDADGGLFPLAFA 861
Query: 461 VVEAECKESWRWFLNHLMRDIGDNGE--KKWVFISDKQKGLLQVFAEDFPGIEHRYCVRH 518
VV+ E ESW WFL+ L + N E + +F+SD QKG++ FP H +C+RH
Sbjct: 862 VVDVENDESWTWFLSELHNALEVNTECMPQIIFLSDGQKGIVDAIRRKFPRSSHAFCMRH 1041
Query: 519 LYANFKQKFGGGTEVRDLLMWASKATYKELWEEKMQEIKKINEAAYWWLMAIPTIHWCRH 578
L N ++F + LL A+ AT + EKM EI++++ A WL W
Sbjct: 1042LSENIGKEFKNSRLIH-LLWSAAYATTINAFREKMAEIEEVSPNASMWLQHFHPSQW--- 1209
Query: 579 AFSYFPKCDV-TMNNLAEAFNCTILVARDKPLITMFEWIRKYLMTRFATLREKSKKWKGN 637
A YF +++ E FN IL A++ P+I + E I+ L T F R KS W
Sbjct: 1210ALVYFEGTRYGHLSSNIEEFNKWILEAQELPIIQVIERIQSKLKTEFDDRRLKSSSWCSV 1389
Query: 638 VMPKPRKRLDREIKYSGNWVATWVAQGLFQVEHAGFEDQFIVDIDKQSCTCNYWELNGIP 697
+ P +R+ I + + + F+V A D IV+I SC+C W+L GIP
Sbjct: 1390LTPSSERRMVEAINRASTYQVLKSDEVEFEVISADRSD--IVNIGSHSCSCRDWQLYGIP 1563
Query: 698 CRHAVACFNDKGLKPENYVHQYYLRATYE--ICYGHMISPINGEN 740
C HAVA Y + + A+Y +C G ++P++ EN
Sbjct: 1564CSHAVAALISSRKDVYAYTAKCFTVASYRDTVCRG--VTPLSLEN 1692
Score = 36.2 bits (82), Expect = 0.060
Identities = 25/82 (30%), Positives = 36/82 (43%), Gaps = 6/82 (7%)
Frame = +3
Query: 728 CYGHMISPINGENKW---PKMSQDDIL---PPEIKRGPGRPKKLRRREPDEPDKKNPTKL 781
CY +P+ G+ +W +DI PP+ +R PGRP+K R D
Sbjct: 1668 CY----TPVPGKLEWRTDESALDNDIAVVRPPKFRRPPGRPEKKRICVEDH--------- 1808
Query: 782 KRGGSSYTCGRCHQKGHNQRKC 803
R + C RC+Q GH + C
Sbjct: 1809 NRDKHTVHCSRCNQTGHYKTTC 1874
>BG451959 homologue to GP|6175165|gb| Mutator-like transposase {Arabidopsis
thaliana}, partial (28%)
Length = 658
Score = 99.4 bits (246), Expect = 6e-21
Identities = 55/166 (33%), Positives = 88/166 (52%), Gaps = 2/166 (1%)
Frame = +2
Query: 362 AVEGDALKQFTLITSYAAELMRVNHENTAAIILRYIPTTLQPRFGCFYMCLEGCKSAFKL 421
A+ G + + L+ Y A + R N + A++ Q F F + G +A
Sbjct: 170 AMRGSFEEGYRLLPQYCAHVKRTNPGSIASVYGNPSDNCFQRLFISFQASIYGLLNA--- 340
Query: 422 ACRPFIGIDGCHLKTKYGGILLIAVGRDPNDQYFPLAFAVVEAECKESWRWFLN--HLMR 479
CRP +G+D +LK+KY G LL+A G D + FPLAF VV+ E ++W WFL+ H +
Sbjct: 341 -CRPLLGLDRIYLKSKYLGTLLLATGFDGDGALFPLAFGVVDEENDDNWMWFLSKLHNLL 517
Query: 480 DIGDNGEKKWVFISDKQKGLLQVFAEDFPGIEHRYCVRHLYANFKQ 525
+I + +SD+Q+G++ +FP H +C+RHL NF++
Sbjct: 518 EINTENMPRLTILSDRQQGIVDGVEANFPTAFHGFCMRHLSDNFRK 655
>CB891859 similar to GP|6175165|gb| Mutator-like transposase {Arabidopsis
thaliana}, partial (27%)
Length = 625
Score = 90.1 bits (222), Expect = 3e-18
Identities = 59/209 (28%), Positives = 102/209 (48%)
Frame = +1
Query: 480 DIGDNGEKKWVFISDKQKGLLQVFAEDFPGIEHRYCVRHLYANFKQKFGGGTEVRDLLMW 539
++ + +SD+Q+G++ +FP H +C+RHL +F+++F T + +LL
Sbjct: 7 EVNTENMPRLTILSDRQQGIVDGVEANFPTAFHGFCMRHLSDSFRKEFNN-TMLVNLLWE 183
Query: 540 ASKATYKELWEEKMQEIKKINEAAYWWLMAIPTIHWCRHAFSYFPKCDVTMNNLAEAFNC 599
A+ +E K+ EI++I++ A +W+ +P W F +T N + EA N
Sbjct: 184 AANCLTIIEFEGKVMEIEEISQDAAYWIRRVPPRLWATAYFEGHRFGHLTAN-IVEALNS 360
Query: 600 TILVARDKPLITMFEWIRKYLMTRFATLREKSKKWKGNVMPKPRKRLDREIKYSGNWVAT 659
IL A P+I M E IR+ LMT F RE S +W ++P + + ++ + +
Sbjct: 361 WILEASGLPIIQMMECIRRQLMTWFNERRETSMQWTSILVPSAERSVAEALERARTYQVL 540
Query: 660 WVAQGLFQVEHAGFEDQFIVDIDKQSCTC 688
+ F+V E IVDI + C C
Sbjct: 541 RANEAEFEV--ISHEGTNIVDIRNRCCLC 621
>AL375902 similar to GP|9759134|dbj mutator-like transposase-like protein
{Arabidopsis thaliana}, partial (20%)
Length = 542
Score = 80.5 bits (197), Expect = 3e-15
Identities = 44/134 (32%), Positives = 63/134 (46%), Gaps = 11/134 (8%)
Frame = +2
Query: 684 QSCTCNYWELNGIPCRHAVACFNDKGLKPENYVHQYYLRATYEICYGHMISPINGENKWP 743
+ C+C W+L G PC HA A G + + +Y + Y MI PI +++W
Sbjct: 8 RECSCRRWQLYGYPCAHAAAALISCGHNAHMFAEPCFTVQSYRMAYSQMIYPIPDKSQWR 187
Query: 744 KMSQD---------DIL--PPEIKRGPGRPKKLRRREPDEPDKKNPTKLKRGGSSYTCGR 792
+ + DI+ PP+I+R PGRPKK R + KR CGR
Sbjct: 188 EHGEGAEGGGGARVDIVIHPPKIRRPPGRPKKKVLRVEN---------FKRPKRVVQCGR 340
Query: 793 CHQKGHNQRKCPLP 806
CH GH+Q+KC +P
Sbjct: 341 CHMLGHSQKKCTMP 382
>BF646328 similar to PIR|F84533|F845 Mutator-like transposase [imported] -
Arabidopsis thaliana, partial (3%)
Length = 636
Score = 57.4 bits (137), Expect(2) = 5e-15
Identities = 34/119 (28%), Positives = 54/119 (44%), Gaps = 3/119 (2%)
Frame = -1
Query: 605 RDKPLITMFEWIRKYLMTRFATLREKSKKWKGNVMPKPRKRLDREIKYSGNWVATW---V 661
R+KP+I + E + + + K + M K + K W ATW +
Sbjct: 630 REKPVIXLLEGYQALYNCEDCQAKADA*KVQW*DMSKDSSFD*EKQKSCRGWKATWHGDM 451
Query: 662 AQGLFQVEHAGFEDQFIVDIDKQSCTCNYWELNGIPCRHAVACFNDKGLKPENYVHQYY 720
F V + +++IV++ +++C C W L GIPC H + C GL EN+V YY
Sbjct: 450 EMNNFNVSNE--TNKYIVNLAQRTCACRKWNLTGIPCAHVIPCIWHNGLADENFVSSYY 280
Score = 42.4 bits (98), Expect(2) = 5e-15
Identities = 18/44 (40%), Positives = 26/44 (58%)
Frame = -3
Query: 729 YGHMISPINGENKWPKMSQDDILPPEIKRGPGRPKKLRRREPDE 772
Y H++ P +G WP + + I P +R GRPKKLR++ DE
Sbjct: 169 YSHIVLPSSGPKLWPVTNTEHINPLAKRRSAGRPKKLRKKANDE 38
>BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (39%)
Length = 1358
Score = 66.6 bits (161), Expect = 4e-11
Identities = 59/193 (30%), Positives = 70/193 (35%), Gaps = 4/193 (2%)
Frame = -2
Query: 743 PKMSQDDILPPEIKRGPGRPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRK 802
P+ S PPE P R R P P P + G + R
Sbjct: 553 PRQSPRPRPPPEPLHNP------RPRPPPRPPPAAPARAPPSGPPAPPTPPATRAPPVRA 392
Query: 803 CPLPPAKKQPTPS---QPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARAR 859
P P + Q TP + + P PP P RA PPPPPA A+R R
Sbjct: 391 SPWCPGQGQETPQGRHTGVQSPVPCPALPHPPGPPPPLDRA----PPPPPAPLPLASRLR 224
Query: 860 HQA-PQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKP 918
+A P PPP P AA A + P PPPP P + P QP+ P S P
Sbjct: 223 PRASPPPPPPPPGPAASAPPRPPPPPPPPP------QRPPPPPPPHTHQPEPPT---SPP 71
Query: 919 AAQPTKKPATKPS 931
QP A PS
Sbjct: 70 RRQPPAPRAPPPS 32
Score = 66.6 bits (161), Expect = 4e-11
Identities = 51/168 (30%), Positives = 61/168 (35%), Gaps = 46/168 (27%)
Frame = -2
Query: 805 LPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAA-------- 856
L P ++ P P P P P P PAA ARA PP PP PA A
Sbjct: 562 LAPPRQSPRPRPPPEPLHNPRPRPPPRPPPAAPARAPPSGPPAPPTPPATRAPPVRASPW 383
Query: 857 ---------RARH-----------------------QAPQPPPASPAAAARARHQA---P 881
+ RH +AP PPPA A+R R +A P
Sbjct: 382 CPGQGQETPQGRHTGVQSPVPCPALPHPPGPPPPLDRAPPPPPAPLPLASRLRPRASPPP 203
Query: 882 QPPPPSPAAAARARHEGEPAARSASQPKKP---AVQPSKPAAQPTKKP 926
PPPP PAA+A R P P P QP P + P ++P
Sbjct: 202 PPPPPGPAASAPPRPPPPPPPPPQRPPPPPPPHTHQPEPPTSPPRRQP 59
Score = 61.6 bits (148), Expect = 1e-09
Identities = 49/132 (37%), Positives = 55/132 (41%), Gaps = 5/132 (3%)
Frame = -1
Query: 804 PLPPAKKQPTPSQPA-AAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQ- 861
P PP P P P+ A ++A PPP P AR R PP PA RA Q
Sbjct: 572 PRPPRPPPPVPPPPSPPRAPPQSEAAPPPPPPPRGARPRAPLRPPSAPDPAGHPRAPRQG 393
Query: 862 -APQPPPASPAAAARARHQAPQPPP-PSPAAAARARHEGEPAARSASQPKKPAVQP-SKP 918
A P P S + RA H+ P+P P P P R PAA P P P SKP
Sbjct: 392 VAVVPGPRS-GNSPRAAHRGPEPRPLPRPPPPTR-----PPAAPGPGPPPAPGPTPLSKP 231
Query: 919 AAQPTKKPATKP 930
A PAT P
Sbjct: 230 PAAEGIPPATAP 195
Score = 60.1 bits (144), Expect = 4e-09
Identities = 44/136 (32%), Positives = 54/136 (39%), Gaps = 10/136 (7%)
Frame = -1
Query: 805 LPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARA-------RHQAPPPPPASPAAAAR 857
LPP + P P + +A QP P+ R R P PPP SP A
Sbjct: 689 LPPPRPPPPPPEWERVPLAKKIGGQPRPALVPLVRVATYPRPPRPPPPVPPPPSPPRAPP 510
Query: 858 ARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEG---EPAARSASQPKKPAVQ 914
AP PPP P AR R P P PA RA +G P RS + P + A +
Sbjct: 509 QSEAAPPPPP--PPRGARPRAPLRPPSAPDPAGHPRAPRQGVAVVPGPRSGNSP-RAAHR 339
Query: 915 PSKPAAQPTKKPATKP 930
+P P P T+P
Sbjct: 338 GPEPRPLPRPPPPTRP 291
Score = 58.5 bits (140), Expect = 1e-08
Identities = 50/157 (31%), Positives = 56/157 (34%), Gaps = 30/157 (19%)
Frame = -1
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAA--------- 854
P PP P PS P A ++A PPP P AR R PP PA
Sbjct: 563 PRPPPPVPPPPSPPRAPP--QSEAAPPPPPPPRGARPRAPLRPPSAPDPAGHPRAPRQGV 390
Query: 855 ----------AARARHQAPQ--------PPPASPAAAARARHQAPQPPPPSPAAAARA-- 894
+ RA H+ P+ PP PAA AP P P S AA
Sbjct: 389 AVVPGPRSGNSPRAAHRGPEPRPLPRPPPPTRPPAAPGPGPPPAPGPTPLSKPPAAEGIP 210
Query: 895 -RHEGEPAARSASQPKKPAVQPSKPAAQPTKKPATKP 930
P AR P PA P+ PAA P PA P
Sbjct: 209 PATAPPPRARRLRPPSAPAPPPTPPAAAP---PAPTP 108
Score = 57.8 bits (138), Expect = 2e-08
Identities = 56/201 (27%), Positives = 71/201 (34%), Gaps = 32/201 (15%)
Frame = -3
Query: 762 PKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLPPAKKQPTPSQ----P 817
P + RR P P P+ G+ +C + PP + PS+ P
Sbjct: 720 PSRAPRRHPPPPPPPPPSSPPGMGTRPSC--------QENWWAAPPGARPSRPSRYIPPP 565
Query: 818 AAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQP--PPASPAAA-- 873
+ A+ AP PP SP+ R R P PPP P A Q P+P PPA P +
Sbjct: 564 TSPPPASPPAPVPPQSPSTI-RGRAPPPAPPPRRPPARPPPAPQRPRPRRPPARPPSGRR 388
Query: 874 --ARAR-----------HQAPQPPPPSPAAAARAR-----------HEGEPAARSASQPK 909
ARA+ +AP P PPSP A R H AA P
Sbjct: 387 RGARAKVRKLPKGGTPGSRAPSPAPPSPTHPAPRRPWTGPPPRPRPHSP*QAACGRGHPP 208
Query: 910 KPAVQPSKPAAQPTKKPATKP 930
+ P P P P P
Sbjct: 207 RHRPPPPGPPPPPPLGPRPPP 145
Score = 53.9 bits (128), Expect = 3e-07
Identities = 51/195 (26%), Positives = 65/195 (33%), Gaps = 23/195 (11%)
Frame = -3
Query: 759 PGRPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPL--PPAKKQPTPSQ 816
P RP + P P + S GR R+ P PPA ++P P +
Sbjct: 597 PSRPSRYIPPPTSPPPASPPAPVPPQSPSTIRGRAPPPAPPPRRPPARPPPAPQRPRPRR 418
Query: 817 PAAAAIATNQAPQPPPSPAAAARAR-----------HQAPPPPPASPAAAARARHQA--- 862
P A +PP ARA+ +AP P P SP A R
Sbjct: 417 PPA---------RPPSGRRRGARAKVRKLPKGGTPGSRAPSPAPPSPTHPAPRRPWTGPP 265
Query: 863 PQPPPASPAAAA-------RARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQP 915
P+P P SP AA R R P PPPP P H P + ++ P
Sbjct: 264 PRPRPHSP*QAACGRGHPPRHRPPPPGPPPPPPLGPRPPPHPPRSGPPRPHPPTRTSLSP 85
Query: 916 SKPAAQPTKKPATKP 930
P + P P
Sbjct: 84 PPPPPAASPPPPAPP 40
Score = 52.4 bits (124), Expect = 8e-07
Identities = 53/199 (26%), Positives = 74/199 (36%), Gaps = 21/199 (10%)
Frame = -3
Query: 753 PEIKRGPGRPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLPPAKKQP 812
P ++R R + R P +++P + S+ H +R P PP P
Sbjct: 834 PPLRRSIVRARAARAVRPPRTAQRHPPRRTLRASAAQ----HPSRAPRRHPPPPPP---P 676
Query: 813 TPSQPAAAAIATNQAPQP-----PPSPAAAARARHQAPP--PPPASPAAAA--------R 857
PS P + T + Q PP + +R+ PP PPPASP A R
Sbjct: 675 PPSSPPG--MGTRPSCQENWWAAPPGARPSRPSRYIPPPTSPPPASPPAPVPPQSPSTIR 502
Query: 858 ARHQAPQPPPASPAAAARARHQAPQP------PPPSPAAAARARHEGEPAARSASQPKKP 911
R P PPP P A Q P+P PP ARA+ P + + P
Sbjct: 501 GRAPPPAPPPRRPPARPPPAPQRPRPRRPPARPPSGRRRGARAKVRKLPKGGTPGS-RAP 325
Query: 912 AVQPSKPAAQPTKKPATKP 930
+ P P ++P T P
Sbjct: 324 SPAPPSPTHPAPRRPWTGP 268
Score = 45.8 bits (107), Expect = 8e-05
Identities = 48/164 (29%), Positives = 63/164 (38%), Gaps = 20/164 (12%)
Frame = -2
Query: 773 PDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLPPAKKQPTPSQPAAAAIATNQAP--QP 830
P +P + C R H P PP + P + A+ T P P
Sbjct: 850 PGPPDPASTPLDRARACCARRPPSPHR----PAPPPQAHPARERRTASLARTPPPPPSSP 683
Query: 831 PPSPAAAARARHQAPPP-----PPA-----------SPAAAARARHQAPQP-PPASPAAA 873
PP+P R + + P PA +PA A R Q+P+P PP P
Sbjct: 682 PPAPLLPPRNGNASLLPRKLVGSPARRSSLSSESLHTPAHLAPPR-QSPRPRPPPEPLHN 506
Query: 874 ARARHQAPQPPP-PSPAAAARARHEGEPAARSASQPKKPAVQPS 916
R P+PPP P PAA ARA G PA + + P V+ S
Sbjct: 505 PR-----PRPPPRPPPAAPARAPPSGPPAPPTPPATRAPPVRAS 389
Score = 42.0 bits (97), Expect = 0.001
Identities = 37/143 (25%), Positives = 51/143 (34%), Gaps = 21/143 (14%)
Frame = -3
Query: 805 LPPAKKQPTPSQPAAAAIATNQAPQPPPS----------PAAAARARHQAPPPPPASPAA 854
+PP ++ ++ A A A + PP P+ A R PPPPP S
Sbjct: 837 IPPLRRSIVRARAARAVRPPRTAQRHPPRRTLRASAAQHPSRAPRRHPPPPPPPPPSSPP 658
Query: 855 AARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKK---- 910
R + A+P A +R PPP SP PA+ A P +
Sbjct: 657 GMGTRPSCQENWWAAPPGARPSRPSRYIPPPTSPP----------PASPPAPVPPQSPST 508
Query: 911 -------PAVQPSKPAAQPTKKP 926
PA P +P A+P P
Sbjct: 507 IRGRAPPPAPPPRRPPARPPPAP 439
Score = 40.8 bits (94), Expect = 0.002
Identities = 44/151 (29%), Positives = 56/151 (36%), Gaps = 18/151 (11%)
Frame = -3
Query: 792 RCHQKGHNQRKCPLPPAKKQPTPSQPAAAAIATNQAPQPPPSPA-----------AAARA 840
R + +G+ Q PLP + + P P +PA+A + P P PA A RA
Sbjct: 1041 RLYTEGYVQC-APLPRSARSPRPHRPASAPSS------PAPLPARDCRRGTLLFRALTRA 883
Query: 841 RHQAPPPPPASPAAAARARHQAPQPPP-------ASPAAAARARHQAPQPPPPSPAAAAR 893
R A SPA PQ PP A A A R A + PP A+
Sbjct: 882 RSTARRRSSTSPA---------PQIPPLRRSIVRARAARAVRPPRTAQRHPPRRTLRASA 730
Query: 894 ARHEGEPAARSASQPKKPAVQPSKPAAQPTK 924
A+H R P P PS P T+
Sbjct: 729 AQHPSRAPRRHPPPPPPP--PPSSPPGMGTR 643
Score = 40.4 bits (93), Expect = 0.003
Identities = 42/144 (29%), Positives = 52/144 (35%), Gaps = 31/144 (21%)
Frame = -1
Query: 818 AAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAA-------RARHQAPQPPPASP 870
AAA ++ + + P A RA P PP S AA RA P PP A+P
Sbjct: 923 AAAVLSCSARSRARDRPRADVRAL----PRPPRSRLYAARSCARVLRAPSALPAPPSATP 756
Query: 871 --AAAARARHQ-------------APQPPPPSP---------AAAARARHEGEPAARSAS 906
A ARA H P+PPPP P + R P R A+
Sbjct: 755 PGAPCARAPHSIPRAHPAATPLLPPPRPPPPPPEWERVPLAKKIGGQPRPALVPLVRVAT 576
Query: 907 QPKKPAVQPSKPAAQPTKKPATKP 930
P+ P +P P P P P
Sbjct: 575 YPRPP--RPPPPVPPPPSPPRAPP 510
Score = 38.5 bits (88), Expect = 0.012
Identities = 40/135 (29%), Positives = 53/135 (38%), Gaps = 19/135 (14%)
Frame = -2
Query: 807 PAKKQPTPS-------QPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPAS--PAAAAR 857
PA ++P PS AA + AP P +R+ A PP S P A AR
Sbjct: 1063 PAPRRPAPSIYGGLCAVRAAPSERAFSAPAPSGVCPVVSRSSTGARLPPRYSLVPRAHAR 884
Query: 858 ARHQAPQ----PPPASPAAAA--RARHQAPQPPP----PSPAAAARARHEGEPAARSASQ 907
+AP P P PA+ RAR + PP P+P A E A+ + +
Sbjct: 883 EIDRAPTFEHFPGPPDPASTPLDRARACCARRPPSPHRPAPPPQAHPARERRTASLARTP 704
Query: 908 PKKPAVQPSKPAAQP 922
P P+ P P P
Sbjct: 703 PPPPSSPPPAPLLPP 659
Score = 35.0 bits (79), Expect = 0.13
Identities = 36/106 (33%), Positives = 42/106 (38%), Gaps = 10/106 (9%)
Frame = -2
Query: 817 PAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAA---RARHQAPQPPPASPAAA 873
PA A AP SPA A +P SP AA R R + A
Sbjct: 1357 PAVDAPPRALAPSRTRSPAVGITAL-SSPFVSSRSPRAARDTLRRRGVGARARRARETHL 1181
Query: 874 ARAR------HQAPQPPPPSPA-AAARARHEGEPAARSASQPKKPA 912
RAR +APQP P PA AA R R + R A P++PA
Sbjct: 1180 PRARIRDRPYSRAPQPAPAPPAHAALRTRRSAPASRRRAPAPRRPA 1043
>TC86766 weakly similar to PIR|T06291|T06291 extensin homolog T9E8.80 -
Arabidopsis thaliana, partial (18%)
Length = 1260
Score = 58.9 bits (141), Expect = 9e-09
Identities = 42/139 (30%), Positives = 46/139 (32%), Gaps = 8/139 (5%)
Frame = +2
Query: 801 RKCPLPPAKKQPTPSQPAAAAIATNQAP----QPPPSPAAAARARHQAPPPPPASPAAAA 856
R P PP P P PA P PPP PA + +PPPPP SP
Sbjct: 23 RSPPPPPPNSPPPPPPPAPVFSPPPPVPYYYSSPPPPPAHSPPPPPXSPPPPPHSPTPPV 202
Query: 857 RARHQAPQPPPASPAAAARARHQAP----QPPPPSPAAAARARHEGEPAARSASQPKKPA 912
P PPP H P PPPPSP P P PA
Sbjct: 203 YPYLSPPPPPPV---------HSPPPPVYSPPPPSPPPCVEPPPPPPPPCVEPPPPSSPA 355
Query: 913 VQPSKPAAQPTKKPATKPS 931
P + P P+ PS
Sbjct: 356 --PHQTPYHPPPSPSPPPS 406
Score = 57.4 bits (137), Expect = 3e-08
Identities = 38/118 (32%), Positives = 46/118 (38%), Gaps = 3/118 (2%)
Frame = +2
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAP 863
P PP P PS P + P PPP P PPPP+SPA HQ P
Sbjct: 245 PPPPVYSPPPPSPPPCV-----EPPPPPPPPCVE--------PPPPSSPAP-----HQTP 370
Query: 864 QPPPASPAAAARARHQAPQPPPP---SPAAAARARHEGEPAARSASQPKKPAVQPSKP 918
PP SP+ + P PPPP SP + + P +S P P + P
Sbjct: 371 YHPPPSPSPPPSPVYAYPSPPPPVYTSPPPSPVYAYPSPPPPVYSSPPPPPVYEGPIP 544
Score = 52.8 bits (125), Expect = 6e-07
Identities = 36/122 (29%), Positives = 42/122 (33%), Gaps = 7/122 (5%)
Frame = +2
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPP----PPASPAAAARAR 859
P PP P P P P PPP H PPP PP SP
Sbjct: 149 PPPPXSPPPPPHSPTPPVYPYLSPPPPPPV--------HSPPPPVYSPPPPSPPPCVEPP 304
Query: 860 HQAPQPPPASPAAAARARHQAPQPPPPSPA---AAARARHEGEPAARSASQPKKPAVQPS 916
P P P ++ A HQ P PPPSP+ + A P ++ P PS
Sbjct: 305 PPPPPPCVEPPPPSSPAPHQTPYHPPPSPSPPPSPVYAYPSPPPPVYTSPPPSPVYAYPS 484
Query: 917 KP 918
P
Sbjct: 485 PP 490
>BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xylostella
granulovirus}, partial (42%)
Length = 909
Score = 58.2 bits (139), Expect = 1e-08
Identities = 47/169 (27%), Positives = 63/169 (36%), Gaps = 3/169 (1%)
Frame = +1
Query: 759 PGRPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLPPAKKQPTPSQPA 818
P RP K + + P + TK ++ R +K N PP + P P P
Sbjct: 28 PKRPPKEKDTQARNPQPEEATKKQK--------RAPEKKKN------PPRGQPPPPPPPP 165
Query: 819 AAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQPPPASPAAAARARH 878
A PP P ++ PPPPP +P Q P PP P R R
Sbjct: 166 PQA--------PPTKPPTRQTPKNNTPPPPPHTPPDPTPP--QPPPQPPKPPHHEKRPRT 315
Query: 879 QAP---QPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPTK 924
P +PPPP+ AR + AR A +P+ P P + TK
Sbjct: 316 PEPPGGRPPPPTTPKQARPTTQERGGARGA-RPRPPPAPPQGTGGEDTK 459
Score = 53.9 bits (128), Expect = 3e-07
Identities = 45/179 (25%), Positives = 59/179 (32%)
Frame = +2
Query: 752 PPEIKRGPGRPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLPPAKKQ 811
PPE P RP+ R +P+ K P K H+ P P
Sbjct: 128 PPEASHPPPRPRPPRPPQPNHQHDK-PQKTTHP-------------HHPHTPPPTPHPPN 265
Query: 812 PTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQPPPASPA 871
P P+ P + P+ P A R R P PPP +AP PPP P
Sbjct: 266 PPPNHPNHPTTRSAHGPRNHPGDAPHRRRRPNKPGPPPRRGGGQG---GRAPAPPPPPPK 436
Query: 872 AAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPTKKPATKP 930
A + P+P P S R + PA + K P P + + P P
Sbjct: 437 A---REGRTPKPDPGSKENHNRTPGKEGPATHGPPEKKTPPPTPKRRGKHTKRAPGGGP 604
Score = 53.1 bits (126), Expect = 5e-07
Identities = 45/163 (27%), Positives = 57/163 (34%), Gaps = 9/163 (5%)
Frame = +3
Query: 773 PDKKNPTKLKRGGSSYTCGRCHQ-----KGHNQRKCPLPPAKKQPTPSQPAAAAIATNQA 827
P + P + RGG + +G R P KK P P Q T +
Sbjct: 414 PPPRPPPRHGRGGHQNQTREAKRTTTEPQGKRDRPRTDPRKKKHPPPHQKEGG--NTQKE 587
Query: 828 PQ----PPPSPAAAARARHQAPPPPPASPAAAARARHQAPQPPPASPAAAARARHQAPQP 883
P+ PP +PA PPPP A R P A ARAR + +
Sbjct: 588 PRGEDPPPHTPAGGGGGPGGPRPPPPKKTARGGEER----------PPAGARARREKTRG 737
Query: 884 PPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPTKKP 926
A A R R + A + K P +P KP QP K P
Sbjct: 738 DQQEAAPAGRPRKKPRGAREREGKKKNPQKKPPKPQQQPPKNP 866
Score = 52.8 bits (125), Expect = 6e-07
Identities = 38/139 (27%), Positives = 50/139 (35%), Gaps = 13/139 (9%)
Frame = +1
Query: 804 PLPPAKKQPTPS--------QPAAAAIATNQAPQPPPSPAAAARARHQAPPP-----PPA 850
P PPA K+P QP A +AP+ +P R + PPP PP
Sbjct: 13 PQPPAPKRPPKEKDTQARNPQPEEATKKQKRAPEKKKNPP---RGQPPPPPPPPPQAPPT 183
Query: 851 SPAAAARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKK 910
P ++ P PPP +P P PP P P HE P P+
Sbjct: 184 KPPTRQTPKNNTPPPPPHTP--------PDPTPPQPPPQPPKPPHHEKRPRT-----PEP 324
Query: 911 PAVQPSKPAAQPTKKPATK 929
P +P P +P T+
Sbjct: 325 PGGRPPPPTTPKQARPTTQ 381
Score = 52.8 bits (125), Expect = 6e-07
Identities = 50/187 (26%), Positives = 68/187 (35%), Gaps = 26/187 (13%)
Frame = +3
Query: 770 PDEPDKKNP--TKLKRGGSSYT--CGRCHQKG----HNQRKCPLPPAKKQPTPSQPAA-- 819
P +P +P T ++G S GR H+K + K P PA P P+ P
Sbjct: 3 PHDPPTPSPQATPQRKGHPSQKPPTGRSHKKTKKSPRKKEKPPPRPATPPPAPAPPGPPN 182
Query: 820 -AAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAP-----QPPPASPAAA 873
TN Q P+ H P PPP + R P PP A A
Sbjct: 183 QTTNTTNPKKQHTPTTPTHPPRPHTPPTPPPTTQTTPPREAPTDPGTTRGTPPTADDAQT 362
Query: 874 ARARHQ--------APQPPPPSPAAAARARHEGE--PAARSASQPKKPAVQPSKPAAQPT 923
+ A H AP PPP P R H+ + A R+ ++P+ + +P P
Sbjct: 363 SPAHHPGEGGGKGGAPPPPPRPPPRHGRGGHQNQTREAKRTTTEPQG---KRDRPRTDPR 533
Query: 924 KKPATKP 930
KK P
Sbjct: 534 KKKHPPP 554
Score = 48.9 bits (115), Expect = 9e-06
Identities = 51/187 (27%), Positives = 63/187 (33%), Gaps = 8/187 (4%)
Frame = +2
Query: 752 PPEIKRGPGRPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLPPAKKQ 811
PP K GR K +P + N T K G +++ PP KK
Sbjct: 422 PPPPKAREGRTPK---PDPGSKENHNRTPGKEGPATHG----------------PPEKKT 544
Query: 812 PTPSQPAAAAIATNQAP---QPPPSPAAAARARHQAPPPPPASPA-----AAARARHQAP 863
P P+ P T +AP PPP P PPPPP + AA R R
Sbjct: 545 PPPT-PKRRGKHTKRAPGGGPPPPHPGRGGGGPGGPPPPPPKKNSERGGRAAPRGRPGTK 721
Query: 864 QPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPT 923
+ P PA + A + P R R G+ PK P KP P
Sbjct: 722 RKNPRRPAGSGPRGAPAKKTP-------RRTRERGKKKKPPKKTPKTTTTTPQKP---PP 871
Query: 924 KKPATKP 930
K T P
Sbjct: 872 HKKNTPP 892
Score = 48.1 bits (113), Expect = 2e-05
Identities = 43/178 (24%), Positives = 59/178 (32%), Gaps = 14/178 (7%)
Frame = +2
Query: 766 RRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLPPAKKQPTPSQPAAAAIATN 825
RRR P++P +GG + K R P K+ P AT+
Sbjct: 344 RRRRPNKPGPPPRRGGGQGGRAPAPPPPPPKAREGRTPKPDPGSKENHNRTPGKEGPATH 523
Query: 826 QAPQ---PPPSPAAAARARHQAP--PPPPASPAAAARARHQAPQPPP---------ASPA 871
P+ PPP+P + +AP PPP P P PPP A+P
Sbjct: 524 GPPEKKTPPPTPKRRGKHTKRAPGGGPPPPHPGRGGGGPGGPPPPPPKKNSERGGRAAPR 703
Query: 872 AAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPTKKPATK 929
+ + P+ P S A A+ + K P P P K P K
Sbjct: 704 GRPGTKRKNPRRPAGSGPRGAPAKKTPRRTRERGKKKKPPKKTPKTTTTTPQKPPPHK 877
Score = 43.9 bits (102), Expect = 3e-04
Identities = 51/205 (24%), Positives = 67/205 (31%), Gaps = 32/205 (15%)
Frame = +2
Query: 753 PEIKRGPGRPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPL-----PP 807
P + P P K R +P+ P++K P K K+ + +RK P PP
Sbjct: 11 PPNPQPPSDPPKKRTPKPETPNRKKPQKNKK------------EPPKKRKTPPEASHPPP 154
Query: 808 AKKQPTPSQP------AAAAIATNQAPQPPPSPAAAARARHQAPPPP------------- 848
+ P P QP + PPP+P H PPP
Sbjct: 155 RPRPPRPPQPNHQHDKPQKTTHPHHPHTPPPTP-------HPPNPPPNHPNHPTTRSAHG 313
Query: 849 PASPAAAARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQP 908
P + A R + P P P +AP PPPP AR +P S
Sbjct: 314 PRNHPGDAPHRRRRPNKPGPPPRRGGGQGGRAPAPPPP--PPKAREGRTPKPDPGSKENH 487
Query: 909 KK------PAVQ--PSKPAAQPTKK 925
+ PA P K PT K
Sbjct: 488 NRTPGKEGPATHGPPEKKTPPPTPK 562
Score = 41.6 bits (96), Expect = 0.001
Identities = 49/191 (25%), Positives = 65/191 (33%), Gaps = 16/191 (8%)
Frame = +1
Query: 752 PPEIKRGPG------RPKKLRRREPDE-PDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCP 804
PP +G G RP K +REP + P ++ + G T +KG +K P
Sbjct: 418 PPAPPQGTGGEDTKTRPGK--QREPQQNPRERGTGHARTPGKKNTPPHTKKKGETHKKSP 591
Query: 805 LPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQ 864
+ P P P P APPP +AP
Sbjct: 592 -------------------GGRTPPPTPRPGGGGARGAPAPPPQKKQREGGKSGPPRAPG 714
Query: 865 PPPASPAAAARARHQAPQPP----PPSPAAAARARHEGE-----PAARSASQPKKPAVQP 915
PAA +R R PP +PAA AR R + + P + + PK P P
Sbjct: 715 HEEKKPAATSRKR-----PPRGAREKNPAAHAREREKKKTPKKNPQNHNNNPPKTP--PP 873
Query: 916 SKPAAQPTKKP 926
K P K P
Sbjct: 874 QKKHPPPPKTP 906
Score = 39.3 bits (90), Expect = 0.007
Identities = 50/230 (21%), Positives = 66/230 (27%), Gaps = 56/230 (24%)
Frame = +1
Query: 753 PEIKRGP--GRPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLPPAKK 810
PE K+ P G+P P P K PT+ Q N P P
Sbjct: 109 PEKKKNPPRGQPPPPPPPPPQAPPTKPPTR--------------QTPKNNTPPPPPHTPP 246
Query: 811 QPTPSQPAAAAIATN------QAPQPP--------------PSPAAAARARHQAPPPPPA 850
PTP QP + P+PP P+ AR P PPPA
Sbjct: 247 DPTPPQPPPQPPKPPHHEKRPRTPEPPGGRPPPPTTPKQARPTTQERGGARGARPRPPPA 426
Query: 851 SPAAA--------------------------ARARHQAPQPPPASPAAAARARHQAPQPP 884
P AR + PP + + P
Sbjct: 427 PPQGTGGEDTKTRPGKQREPQQNPRERGTGHARTPGKKNTPPHTKKKGETHKKSPGGRTP 606
Query: 885 PPSP--------AAAARARHEGEPAARSASQPKKPAVQPSKPAAQPTKKP 926
PP+P A A + + + P+ P + KPAA K+P
Sbjct: 607 PPTPRPGGGGARGAPAPPPQKKQREGGKSGPPRAPGHEEKKPAATSRKRP 756
>TC85251 homologue to PIR|JQ2128|JQ2128 metallothionein - soybean, partial
(93%)
Length = 1554
Score = 57.0 bits (136), Expect = 3e-08
Identities = 42/133 (31%), Positives = 53/133 (39%), Gaps = 11/133 (8%)
Frame = -2
Query: 810 KQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPP---PASPAAAARARHQAPQPP 866
K P P P ++ ++T P P P P + Q PP P A+P A QAP P
Sbjct: 668 KVPPPQPPKSSPVSTPTLPPPLPPPPKISPTPVQTPPAPAPVKATPVPAPAPAKQAPTPA 489
Query: 867 PAS--PAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQP------KKPAVQPSKP 918
PA+ P A +AP P P S + R RH + A P K P P+
Sbjct: 488 PATSPPIPAPTPAIEAPVPAPES-SKPKRRRHRPKHRRHQAPAPAPTVIHKSPPAPPTDT 312
Query: 919 AAQPTKKPATKPS 931
A PA PS
Sbjct: 311 TADSDTAPAPAPS 273
Score = 52.0 bits (123), Expect = 1e-06
Identities = 41/114 (35%), Positives = 46/114 (39%), Gaps = 16/114 (14%)
Frame = -2
Query: 804 PLPPAKK-QPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPAS--PAAAARARH 860
PLPP K PTP Q A P P P+PA QAP P PA+ P A
Sbjct: 608 PLPPPPKISPTPVQTPPAPAPVKATPVPAPAPAK------QAPTPAPATSPPIPAPTPAI 447
Query: 861 QAPQPPPASPAAAARA------RHQAPQP-------PPPSPAAAARARHEGEPA 901
+AP P P S R RHQAP P PP+P A + PA
Sbjct: 446 EAPVPAPESSKPKRRRHRPKHRRHQAPAPAPTVIHKSPPAPPTDTTADSDTAPA 285
Score = 50.4 bits (119), Expect = 3e-06
Identities = 37/127 (29%), Positives = 50/127 (39%)
Frame = -2
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAP 863
P P P +QP + QPP + A ++ P P +PA++ + P
Sbjct: 821 PTTPTAITPVTTQPPTVVASPPITSQPPVT--VAPKSAPVTSPAPKIAPASSPKV--PPP 654
Query: 864 QPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPT 923
QPP +SP + P PP SP + PA PA P+K A PT
Sbjct: 653 QPPKSSPVSTPTLPPPLPPPPKISPTPV-----QTPPAPAPVKATPVPAPAPAKQA--PT 495
Query: 924 KKPATKP 930
PAT P
Sbjct: 494 PAPATSP 474
Score = 50.1 bits (118), Expect = 4e-06
Identities = 39/123 (31%), Positives = 52/123 (41%), Gaps = 4/123 (3%)
Frame = -2
Query: 812 PTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPP--ASPAAAARARHQAPQPPPAS 869
P SQP +A AP P+P A + + PPP P +SP + P PP S
Sbjct: 758 PITSQPPVT-VAPKSAPVTSPAPKIAPASSPKVPPPQPPKSSPVSTPTLPPPLPPPPKIS 582
Query: 870 PAAAARARHQAP--QPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPTKKPA 927
P AP P P+PA A +A P A+ P PA P+ A P + +
Sbjct: 581 PTPVQTPPAPAPVKATPVPAPAPAKQA-----PTPAPATSPPIPAPTPAIEAPVPAPE-S 420
Query: 928 TKP 930
+KP
Sbjct: 419 SKP 411
Score = 49.7 bits (117), Expect = 5e-06
Identities = 45/150 (30%), Positives = 57/150 (38%), Gaps = 24/150 (16%)
Frame = -2
Query: 806 PPAKKQPTPSQPAAAAIATNQAPQ---------PPP-----SPAAAARARHQAPPPPPAS 851
PP QP + +A T+ AP+ PPP SP + PPPP S
Sbjct: 761 PPITSQPPVTVAPKSAPVTSPAPKIAPASSPKVPPPQPPKSSPVSTPTLPPPLPPPPKIS 582
Query: 852 PAAAARARHQAPQPPPASPAAAARARHQAPQPPPPS--PAAAARARHEGEPAARSASQPK 909
P AP P A+P A QAP P P + P A E A +S+PK
Sbjct: 581 PTPVQTP--PAPAPVKATPVPAPAPAKQAPTPAPATSPPIPAPTPAIEAPVPAPESSKPK 408
Query: 910 KPAVQP-----SKPAAQPT---KKPATKPS 931
+ +P PA PT K P P+
Sbjct: 407 RRRHRPKHRRHQAPAPAPTVIHKSPPAPPT 318
Score = 39.7 bits (91), Expect = 0.005
Identities = 27/96 (28%), Positives = 37/96 (38%)
Frame = -2
Query: 801 RKCPLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARH 860
++ P P P S P A +AP P P + R RH+ +PA A H
Sbjct: 509 KQAPTPA----PATSPPIPAPTPAIEAPVPAPESSKPKRRRHRPKHRRHQAPAPAPTVIH 342
Query: 861 QAPQPPPASPAAAARARHQAPQPPPPSPAAAARARH 896
++P PP A + AP P P A + H
Sbjct: 341 KSPPAPPTDTTADS---DTAPAPAPSFNLNGAPSNH 243
>TC87476 homologue to PIR|T11622|T11622 extensin class 1 precursor - cowpea,
partial (48%)
Length = 939
Score = 56.6 bits (135), Expect = 4e-08
Identities = 37/124 (29%), Positives = 56/124 (44%), Gaps = 3/124 (2%)
Frame = +3
Query: 798 HNQRKCPLPPAK---KQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAA 854
H++ P PP K P P P+ ++P PPPSP+ +++PPPP ASP
Sbjct: 360 HHRPPSPSPPPPYVYKSPPPPSPSPPPPYVYKSP-PPPSPSPPPPYVYKSPPPPSASPPP 536
Query: 855 AARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQ 914
+ PPP SP+ +++P PPPSP+ ++ P + P P +
Sbjct: 537 P----YYYKSPPPPSPSPPPPYGYKSP--PPPSPSPPPPYIYKSPPPPSPSPPPYHPYLY 698
Query: 915 PSKP 918
S P
Sbjct: 699 NSPP 710
Score = 49.7 bits (117), Expect = 5e-06
Identities = 35/124 (28%), Positives = 52/124 (41%), Gaps = 1/124 (0%)
Frame = +2
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPP-PPASPAAAARARHQA 862
P P K P P P+ ++P PPPSP+ +++PPP P+ P +
Sbjct: 2 PPPYIYKSPPPPSPSPPPPYVYKSP-PPPSPSPPPPYVYKSPPPPSPSPPPP-----YVY 163
Query: 863 PQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQP 922
PPP +P+ +++P PP PSP + P + P P V S P P
Sbjct: 164 KSPPPPTPSPPPPYIYKSPPPPSPSPPPPYVYKSPPPP----SPSPPPPYVYKSPPPPSP 331
Query: 923 TKKP 926
+ P
Sbjct: 332 SPPP 343
Score = 48.1 bits (113), Expect = 2e-05
Identities = 38/132 (28%), Positives = 53/132 (39%), Gaps = 4/132 (3%)
Frame = +2
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPP-PPASPAAAARARHQA 862
P P K P P P+ ++P PPPSP+ +++PPP P+ P +
Sbjct: 50 PPPYVYKSPPPPSPSPPPPYVYKSP-PPPSPSPPPPYVYKSPPPPTPSPPPP-----YIY 211
Query: 863 PQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKP---A 919
PPP SP+ +++P PP PSP P P P+ P P
Sbjct: 212 KSPPPPSPSPPPPYVYKSPPPPSPSP-----------PPPYVYKSPPPPSPSPPPPYIYK 358
Query: 920 AQPTKKPATKPS 931
+ PT P T S
Sbjct: 359 SPPTTIPFTTTS 394
Score = 45.8 bits (107), Expect = 8e-05
Identities = 32/117 (27%), Positives = 46/117 (38%), Gaps = 12/117 (10%)
Frame = +3
Query: 822 IATNQAPQPPPSPAAAARARHQAPP------------PPPASPAAAARARHQAPQPPPAS 869
I+T+ P PSP + PP PPP SP+ +++P PP AS
Sbjct: 348 ISTSHHRPPSPSPPPPYVYKSPPPPSPSPPPPYVYKSPPPPSPSPPPPYVYKSPPPPSAS 527
Query: 870 PAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPTKKP 926
P + PPPPSP+ ++ P + P P + S P P+ P
Sbjct: 528 PPPP----YYYKSPPPPSPSPPPPYGYKSPPP--PSPSPPPPYIYKSPPPPSPSPPP 680
Score = 42.4 bits (98), Expect = 8e-04
Identities = 26/83 (31%), Positives = 36/83 (43%)
Frame = +3
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAP 863
P P K P P + ++P PPPSP+ +++PPPP SP +
Sbjct: 483 PPPYVYKSPPPPSASPPPPYYYKSP-PPPSPSPPPPYGYKSPPPPSPSPPPP----YIYK 647
Query: 864 QPPPASPAAAARARHQAPQPPPP 886
PPP SP+ + PPPP
Sbjct: 648 SPPPPSPSPPPYHPYLYNSPPPP 716
Score = 31.6 bits (70), Expect = 1.5
Identities = 18/76 (23%), Positives = 25/76 (32%), Gaps = 3/76 (3%)
Frame = +3
Query: 858 ARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSK 917
+ H+ P P P P +P PPPP + P P P+ P
Sbjct: 357 SHHRPPSPSPPPPYVYKSPPPPSPSPPPPYVYKSPPPPSPSPPPPYVYKSPPPPSASPPP 536
Query: 918 P---AAQPTKKPATKP 930
P + P P+ P
Sbjct: 537 PYYYKSPPPPSPSPPP 584
>TC76311 homologue to GP|3204132|emb|CAA07235.1 extensin {Cicer arietinum},
partial (77%)
Length = 973
Score = 56.6 bits (135), Expect = 4e-08
Identities = 35/105 (33%), Positives = 47/105 (44%)
Frame = +2
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAP 863
P P K P P P+ Q+P PPPSP +++PPPP ASP + +P
Sbjct: 116 PPPYYYKSPPPPSPSPPPPYYYQSP-PPPSPTPHTPYHYKSPPPPTASPPPPYH--YVSP 286
Query: 864 QPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQP 908
PP +SP + PPPPSPA A ++ P + P
Sbjct: 287 PPPTSSPPP-----YHYTSPPPPSPAPAPTYIYKSPPPPMKSPPP 406
Score = 50.1 bits (118), Expect = 4e-06
Identities = 33/119 (27%), Positives = 47/119 (38%)
Frame = +2
Query: 812 PTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQPPPASPA 871
P+PS P + P P P P ++ PPP P+ P + PPP SP+
Sbjct: 5 PSPSPPPPYYYHSPPPPSPSPPPPYYYKS---PPPPSPSPPPP-----YYYKSPPPPSPS 160
Query: 872 AAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPTKKPATKP 930
+Q+ PPPPSP ++ P ++ P V P P + P T P
Sbjct: 161 PPPPYYYQS--PPPPSPTPHTPYHYKSPPPPTASPPPPYHYVSPPPPTSSPPPYHYTSP 331
Score = 49.3 bits (116), Expect = 7e-06
Identities = 35/119 (29%), Positives = 51/119 (42%)
Frame = +2
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAP 863
P P P P P+ ++P PPPSP+ +++PPPP SP+ +Q+P
Sbjct: 20 PPPYYYHSPPPPSPSPPPPYYYKSP-PPPSPSPPPPYYYKSPPPP--SPSPPPPYYYQSP 190
Query: 864 QPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQP 922
PP SP +++P PP SP H P ++S P P P+ P
Sbjct: 191 PPP--SPTPHTPYHYKSPPPPTASPPP---PYHYVSPPPPTSSPPPYHYTSPPPPSPAP 352
Score = 47.8 bits (112), Expect = 2e-05
Identities = 29/93 (31%), Positives = 38/93 (40%), Gaps = 6/93 (6%)
Frame = +2
Query: 802 KCPLPPAKKQP------TPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAA 855
K P PP+ P +P P+ PPP A+ H PPPP S
Sbjct: 134 KSPPPPSPSPPPPYYYQSPPPPSPTPHTPYHYKSPPPPTASPPPPYHYVSPPPPTS---- 301
Query: 856 ARARHQAPQPPPASPAAAARARHQAPQPPPPSP 888
+ + PPP SPA A +++P PP SP
Sbjct: 302 SPPPYHYTSPPPPSPAPAPTYIYKSPPPPMKSP 400
Score = 43.9 bits (102), Expect = 3e-04
Identities = 29/88 (32%), Positives = 37/88 (41%), Gaps = 5/88 (5%)
Frame = +2
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQP-----PPSPAAAARARHQAPPPPPASPAAAARA 858
P PP+ TP + T P P PP P ++ H PPPP SPA A
Sbjct: 188 PPPPSPTPHTPYHYKSPPPPTASPPPPYHYVSPPPPTSSPPPYHYTSPPPP-SPAPAPTY 364
Query: 859 RHQAPQPPPASPAAAARARHQAPQPPPP 886
+++P PP SP + PPPP
Sbjct: 365 IYKSPPPPMKSPPPPV---YIYASPPPP 439
Score = 40.4 bits (93), Expect = 0.003
Identities = 24/71 (33%), Positives = 31/71 (42%), Gaps = 6/71 (8%)
Frame = +2
Query: 802 KCPLPPAKKQPTPSQPAAAAIATNQAPQ------PPPSPAAAARARHQAPPPPPASPAAA 855
K P PP P P + T+ P PPPSPA A +++PPPP SP
Sbjct: 230 KSPPPPTASPPPPYHYVSPPPPTSSPPPYHYTSPPPPSPAPAPTYIYKSPPPPMKSPPPP 409
Query: 856 ARARHQAPQPP 866
+ +P PP
Sbjct: 410 VYI-YASPPPP 439
>BQ141932 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (30%)
Length = 1338
Score = 56.2 bits (134), Expect = 6e-08
Identities = 42/138 (30%), Positives = 56/138 (40%), Gaps = 11/138 (7%)
Frame = -1
Query: 804 PLPPAKKQP-TPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQA 862
P PPA + P T ++P + A +PPP P A + + PP PP SP R R
Sbjct: 561 PNPPASQSPATLNRPTSHTPACPPPSRPPPHPRAPSLSP-PPPPHPPTSPPRPGRPRSST 385
Query: 863 PQPPP----ASPAAAAR----ARHQAPQPPPPSPAAAARARHEGEPAA--RSASQPKKPA 912
PPP SP+A P PPPP+P ++ P P +P+
Sbjct: 384 LAPPPHRQFHSPSALCTHCGIPPPAVPPPPPPTPPPSSPPALVSPPPRPFLFPPPPNQPS 205
Query: 913 VQPSKPAAQPTKKPATKP 930
PS P P P +P
Sbjct: 204 PSPSPPPPPPLPPPPPQP 151
Score = 48.1 bits (113), Expect = 2e-05
Identities = 45/177 (25%), Positives = 61/177 (34%), Gaps = 13/177 (7%)
Frame = -1
Query: 758 GPGRPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLPPAKKQPTPSQP 817
G G P+ P ++P L R S + CP PP++ P P P
Sbjct: 597 GTGSAPPAIHHHPNPPASQSPATLNRPTS------------HTPACP-PPSRPPPHPRAP 457
Query: 818 AAAAIATNQAPQPPPSPAAAARARHQAPPPPPA----SPAAA---------ARARHQAPQ 864
+ ++ P PP SP R R PPP SP+A A P
Sbjct: 456 S---LSPPPPPHPPTSPPRPGRPRSSTLAPPPHRQFHSPSALCTHCGIPPPAVPPPPPPT 286
Query: 865 PPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQ 921
PPP+SP A PPP P ++ P+ P P P P ++
Sbjct: 285 PPPSSPPALVS--------PPPRPFLFPPPPNQPSPSPSPPPPPPLPPPPPQPPQSR 139
Score = 45.8 bits (107), Expect = 8e-05
Identities = 39/155 (25%), Positives = 55/155 (35%), Gaps = 19/155 (12%)
Frame = -1
Query: 796 KGHNQRKCP-LPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAA 854
K H + P + P+ P +P + + +A H PP SPA
Sbjct: 708 KAHRSPRPPHVGPSLAHPARPRPGRVLLVMQYPTSFLGTGSAPPAIHHHPNPPASQSPAT 529
Query: 855 AARARHQAPQ-PPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQP----- 908
R P PPP+ P RA +P PPP P + R G P + + + P
Sbjct: 528 LNRPTSHTPACPPPSRPPPHPRAPSLSPPPPPHPPTSPPR---PGRPRSSTLAPPPHRQF 358
Query: 909 ------------KKPAVQPSKPAAQPTKKPATKPS 931
PAV P P PT P++ P+
Sbjct: 357 HSPSALCTHCGIPPPAVPPPPP---PTPPPSSPPA 262
Score = 45.8 bits (107), Expect = 8e-05
Identities = 45/175 (25%), Positives = 62/175 (34%), Gaps = 21/175 (12%)
Frame = -2
Query: 778 PTKLKRGGSSYTCGRCHQKGHNQRKCPLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAA 837
P RGG ++C + + P PP+ TP P T+ P P+P
Sbjct: 656 PPARVRGGFYWSCNTLLRFW--EPAAPHPPSTTTLTPQHPNPPPP*TDPPATPLPAPPHP 483
Query: 838 AR----ARHQAPP-------PPPASPAA------AARARHQAPQPPPASPAAAARARHQA 880
R AR ++PP PPP +P A R + PPP P A
Sbjct: 482 VRPPTHARLRSPPLRPPTLRPPPPAPVAHGHRPSPPRRIGSSTPPPPFVPIAGFPPPPSP 303
Query: 881 PQPPP----PSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPTKKPATKPS 931
P PPP P P ++ H + P P P P + P +P P+
Sbjct: 302 PPPPPLPLHPPPQLSSLPPHAPSSFLLPLTSPPPPPPPPHPPPSPP--RPHNHPN 144
Score = 43.5 bits (101), Expect = 4e-04
Identities = 29/98 (29%), Positives = 35/98 (35%)
Frame = -3
Query: 805 LPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQ 864
LPP + PT P A + P P P + R PPPP + +R P
Sbjct: 421 LPPPPRSPTVIDPRPPAASAVPLPLRPLYPLRDSPPRRPPPPPPHSPSILPPSSRLSPPT 242
Query: 865 PPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAA 902
P P S + P P PP P A PAA
Sbjct: 241 PLPLSSSP*PALPLPLPPPTPPPPPPAPTTTPI*APAA 128
Score = 40.4 bits (93), Expect = 0.003
Identities = 37/143 (25%), Positives = 48/143 (32%), Gaps = 13/143 (9%)
Frame = -3
Query: 798 HNQRKCPLPPAKKQPTPSQPAA---AAIATNQAPQPPPSPAAAARARHQAPPPPPASPAA 854
H C PP P P++ A +A + P PP SP P PPA+ A
Sbjct: 517 HQPHPCLPPPIPSAPPPTRAFALPPSAPPPSDLPPPPRSPTVI-------DPRPPAASAV 359
Query: 855 AARAR----------HQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARS 904
R + P PPP SP+ + +P P P ++ PA
Sbjct: 358 PLPLRPLYPLRDSPPRRPPPPPPHSPSILPPSSRLSPPTPLPLSSSP*-------PALPL 200
Query: 905 ASQPKKPAVQPSKPAAQPTKKPA 927
P P P P P PA
Sbjct: 199 PLPPPTPPPPPPAPTTTPI*APA 131
Score = 40.0 bits (92), Expect = 0.004
Identities = 44/186 (23%), Positives = 61/186 (32%), Gaps = 27/186 (14%)
Frame = -3
Query: 772 EPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLPPAKKQPTPSQPAAAAIATNQAPQPP 831
+P + P +S C + R+C + P P I+ + +PP
Sbjct: 820 DPPRSTPHSATLPPASTHTRSCRRVSAIARRCTRRTREGSPFTPPPPRRTISGSS--RPP 647
Query: 832 PSPAAAAR--------------ARHQAPPPPPASPAAAARARHQAPQP--PPASPAA--A 873
S A + RH PP PP+ P P P PP P+A
Sbjct: 646 ASGAGSTGHAIPYFVFGNRQRPTRHPPPP*PPSIPIPRHPEPTHQPHPCLPPPIPSAPPP 467
Query: 874 ARARHQAPQPPPPS----PAAAARARHEGEPAARSASQPKKPAV-----QPSKPAAQPTK 924
RA P PPPS P + PAA + P +P P +P P
Sbjct: 466 TRAFALPPSAPPPSDLPPPPRSPTVIDPRPPAASAVPLPLRPLYPLRDSPPRRPPPPPPH 287
Query: 925 KPATKP 930
P+ P
Sbjct: 286 SPSILP 269
Score = 39.7 bits (91), Expect = 0.005
Identities = 30/110 (27%), Positives = 37/110 (33%)
Frame = -1
Query: 752 PPEIKRGPGRPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHNQRKCPLPPAKKQ 811
PP PGRP+ P +P+ L C C G P PP
Sbjct: 429 PPTSPPRPGRPRSSTLAPPPHRQFHSPSAL--------CTHC---GIPPPAVPPPPPPTP 283
Query: 812 PTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQ 861
P S PA + PPP + PPP P P ++RHQ
Sbjct: 282 PPSSPPALVSPPPRPFLFPPPPNQPSPSPSPPPPPPLPPPPPQPPQSRHQ 133
Score = 32.7 bits (73), Expect = 0.66
Identities = 36/144 (25%), Positives = 45/144 (31%), Gaps = 28/144 (19%)
Frame = -1
Query: 801 RKCPLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAA-------------RARH-QAPP 846
R P PP T + +++ AP+P P P AA RH P
Sbjct: 1098 RHRPYPPLTSPHTARYSPSLSLSHYPAPRPVPRPPRAAVSILHLPLPTATTHTRHTHTPL 919
Query: 847 PPPASPAAAARARHQAPQPP-----PASPAAAARARHQAPQPPPPSPAAAARARHEGEPA 901
P ++P PP P S R Q P P P RA P
Sbjct: 918 SPRSAPLHLLAPTFTTNTPPLIATRPRSTDITPTLRGQPPTPRPYPQPLRTRAHVGAYPR 739
Query: 902 ARSAS---------QPKKPAVQPS 916
R A+ P+ P V PS
Sbjct: 738 LRDAAHAVLVKAHRSPRPPHVGPS 667
Score = 30.8 bits (68), Expect = 2.5
Identities = 31/107 (28%), Positives = 37/107 (33%), Gaps = 4/107 (3%)
Frame = -3
Query: 794 HQKGHNQRKCPLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPA 853
H + H R PLP P+ A+ T P +A P PPASP
Sbjct: 1138 HARSHIIRVPPLPSPPTIPSSHLAPHRALLTLSLPL----------TLSRASPRPPASP- 992
Query: 854 AAARARHQAPQPPPASPAAAARARHQA-PQPP---PPSPAAAARARH 896
R PP+ H PQPP PPSP A +H
Sbjct: 991 -----RRCLDTTPPSPHRHYTHTTHSHPPQPPLRSPPSPRADFYDQH 866
Score = 29.6 bits (65), Expect = 5.6
Identities = 32/115 (27%), Positives = 38/115 (32%), Gaps = 3/115 (2%)
Frame = -2
Query: 812 PTPSQPAAAAIATNQAPQPPPSPAAAARARHQAP--PPPPASPAAAARARHQAPQPPPAS 869
P P P A T +P+P P H P P P PA +R PP +
Sbjct: 1118 PRPPPPLATD-HTLLSPRPTPRATHPLSPSHTIPRLAPSPGLPAPLSRYYTSLSPPPLHT 942
Query: 870 PAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKP-AVQPSKPAAQPT 923
AP P S H P AR+ +P AV P P PT
Sbjct: 941 HDTLTPPSAPAPLPSISSRRLLRPTLHLSSPPARALPILHRPSAVNP--PLRDPT 783
>BI271388 similar to GP|10177186|db mutator-like transposase-like protein
{Arabidopsis thaliana}, partial (26%)
Length = 711
Score = 56.2 bits (134), Expect = 6e-08
Identities = 31/83 (37%), Positives = 44/83 (52%), Gaps = 2/83 (2%)
Frame = +2
Query: 271 FASTEEFKKAILAHSVMNGKAVIYKKNDKNRVRFACKQEECRFIAYCANIKGTSTYRIKT 330
F S EF++A+ +S+ +G A YKKND +RV CK + C + Y + + T IK
Sbjct: 149 FNSFSEFREALHKYSIAHGFAYRYKKNDSHRVTVKCKSQGCPWRIYASKLSTTQLICIKK 328
Query: 331 LNPDHTC-GRTFK-NRAATAKWI 351
+ DHTC G K AT W+
Sbjct: 329 MTRDHTCEGSAVKAGYRATRGWV 397
>BQ751262 similar to GP|6715100|gb|A versicolorin B synthase {Aspergillus
parasiticus}, partial (11%)
Length = 710
Score = 55.8 bits (133), Expect = 7e-08
Identities = 42/132 (31%), Positives = 60/132 (44%), Gaps = 17/132 (12%)
Frame = +3
Query: 809 KKQPTPSQPAAAAIATNQAPQPPPS-------PAAAARARH--------QAPPPPPASPA 853
+ PTP A +T Q P PPP+ P +R+ H A P P S A
Sbjct: 261 RDSPTP------APSTTQMPSPPPAGLLRSRMPTMPSRSAHISSPLSTRSASPRPRTSTA 422
Query: 854 AAARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKP-- 911
++ AR AP P P +A+RAR ++ P PS A +R+ P S++ K+P
Sbjct: 423 ESSWARSTAPTPSPQRRKSASRARRRS--SPKPSAARTSRSTPSPSPRRSSSTATKRPPA 596
Query: 912 AVQPSKPAAQPT 923
+ P PA+ PT
Sbjct: 597 SSSPPTPASPPT 632
Score = 50.8 bits (120), Expect = 2e-06
Identities = 43/130 (33%), Positives = 63/130 (48%), Gaps = 8/130 (6%)
Frame = +3
Query: 806 PPA----KKQPT-PSQPA--AAAIATNQA-PQPPPSPAAAARARHQAPPPPPASPAAAAR 857
PPA + PT PS+ A ++ ++T A P+P S A ++ AR AP P P +A+R
Sbjct: 309 PPAGLLRSRMPTMPSRSAHISSPLSTRSASPRPRTSTAESSWARSTAPTPSPQRRKSASR 488
Query: 858 ARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSK 917
AR ++ P +AAR P P P ++ A R PA+ S P P PS+
Sbjct: 489 ARRRSSPKP-----SAARTSRSTPSPSPRRSSSTATKR---PPASSSPPTPASPPT-PSR 641
Query: 918 PAAQPTKKPA 927
A + + PA
Sbjct: 642 RARRSSSPPA 671
Score = 46.6 bits (109), Expect = 4e-05
Identities = 36/113 (31%), Positives = 49/113 (42%), Gaps = 5/113 (4%)
Frame = +3
Query: 804 PLPPAKKQPTP-SQPAAAAIATNQAPQPPPSPA-AAARARHQAPPPPPASPAAAARARHQ 861
PL P P + A ++ A + AP P P +A+RAR ++ P P +AAR
Sbjct: 375 PLSTRSASPRPRTSTAESSWARSTAPTPSPQRRKSASRARRRSSPKP-----SAARTSRS 539
Query: 862 APQPPPASPAAAARARHQAPQPPP---PSPAAAARARHEGEPAARSASQPKKP 911
P P P ++ A R A PP P + RAR P A S+P P
Sbjct: 540 TPSPSPRRSSSTATKRPPASSSPPTPASPPTPSRRARRSSSPPA--PSEPPAP 692
Score = 39.3 bits (90), Expect = 0.007
Identities = 21/75 (28%), Positives = 35/75 (46%), Gaps = 2/75 (2%)
Frame = +3
Query: 800 QRKCPLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARA--RHQAPPPPPASPAAAAR 857
QR+ A+++ +P AA + +P P S + A + +PP P + P + R
Sbjct: 465 QRRKSASRARRRSSPKPSAARTSRSTPSPSPRRSSSTATKRPPASSSPPTPASPPTPSRR 644
Query: 858 ARHQAPQPPPASPAA 872
AR + P P+ P A
Sbjct: 645 ARRSSSPPAPSEPPA 689
Score = 33.1 bits (74), Expect = 0.51
Identities = 21/80 (26%), Positives = 34/80 (42%), Gaps = 3/80 (3%)
Frame = +3
Query: 778 PTKLKRGGSSYTCGRCHQK---GHNQRKCPLPPAKKQPTPSQPAAAAIATNQAPQPPPSP 834
P+ +R +S R K R P P ++ + + A ++ P PP+P
Sbjct: 456 PSPQRRKSASRARRRSSPKPSAARTSRSTPSPSPRRSSSTATKRPPASSSPPTPASPPTP 635
Query: 835 AAAARARHQAPPPPPASPAA 854
+ RAR + PP P+ P A
Sbjct: 636 --SRRARRSSSPPAPSEPPA 689
>TC85985 similar to GP|5139695|dbj|BAA81686.1 expressed in cucumber
hypocotyls {Cucumis sativus}, partial (42%)
Length = 892
Score = 55.5 bits (132), Expect = 1e-07
Identities = 44/131 (33%), Positives = 55/131 (41%), Gaps = 8/131 (6%)
Frame = +3
Query: 807 PAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQPP 866
P PT + P+AA +A AP P SPA A + P P + AP P
Sbjct: 138 PTTSPPTVTTPSAAPVA---APTKPKSPAPVASPKSSPPASSPTAATVTPAVSPAAPVPV 308
Query: 867 PASPAAAARARHQAPQPPPPSPAAA--ARARHEGEPAARSASQP---KKPAVQPSK--PA 919
SPAA++ PP P+P ++ A P S P PAV PS PA
Sbjct: 309 AKSPAASSPVVAPVSTPPKPAPVSSPPAPVPVSSPPTPVPVSSPPTASTPAVTPSAEVPA 488
Query: 920 AQPTK-KPATK 929
A P+K K TK
Sbjct: 489 AAPSKSKKKTK 521
Score = 41.6 bits (96), Expect = 0.001
Identities = 35/131 (26%), Positives = 56/131 (42%), Gaps = 10/131 (7%)
Frame = +3
Query: 802 KCPLPPA--KKQPTPSQPAAAAIATNQAPQPP----PSPAAAARARHQAPPPPPASPAAA 855
K P P A K P S P AA + +P P SPAA++ PP +P ++
Sbjct: 204 KSPAPVASPKSSPPASSPTAATVTPAVSPAAPVPVAKSPAASSPVVAPVSTPPKPAPVSS 383
Query: 856 ARARHQAPQPP-PASPAAAARARHQAPQPPPPSPAAA-ARARHEGEPAARSASQPKKPAV 913
A PP P ++ A A P PAAA ++++ + + + ++ PA+
Sbjct: 384 PPAPVPVSSPPTPVPVSSPPTASTPAVTPSAEVPAAAPSKSKKKTKKGKKHSAPAPSPAL 563
Query: 914 Q--PSKPAAQP 922
+ P+ P P
Sbjct: 564 EGPPAPPVGAP 596
Score = 41.2 bits (95), Expect = 0.002
Identities = 30/116 (25%), Positives = 41/116 (34%), Gaps = 16/116 (13%)
Frame = +3
Query: 807 PAKKQPTPSQPAAAAIATNQAPQP---PPSPAAAARARHQAP---PPPPASPAAAARARH 860
P K P S P A ++T P P PP+P + P PP ++PA A
Sbjct: 303 PVAKSPAASSPVVAPVSTPPKPAPVSSPPAPVPVSSPPTPVPVSSPPTASTPAVTPSAEV 482
Query: 861 QAPQPPPASPAAAARARHQAPQPPPP----------SPAAAARARHEGEPAARSAS 906
A P + +H AP P P +P + A G +A S
Sbjct: 483 PAAAPSKSKKKTKKGKKHSAPAPSPALEGPPAPPVGAPGPSLDASSPGPASAADES 650
Score = 32.7 bits (73), Expect = 0.66
Identities = 29/85 (34%), Positives = 35/85 (41%), Gaps = 17/85 (20%)
Frame = +3
Query: 804 PLP-PAKKQPTP----SQPAAAAIA-TNQAPQPPPSPAAAAR-----ARHQAPPP----- 847
P P P PTP S P A+ A T A P +P+ + + +H AP P
Sbjct: 387 PAPVPVSSPPTPVPVSSPPTASTPAVTPSAEVPAAAPSKSKKKTKKGKKHSAPAPSPALE 566
Query: 848 -PPASPAAAARARHQAPQPPPASPA 871
PPA P A A P PAS A
Sbjct: 567 GPPAPPVGAPGPSLDASSPGPASAA 641
>TC87439 weakly similar to GP|5139695|dbj|BAA81686.1 expressed in cucumber
hypocotyls {Cucumis sativus}, partial (57%)
Length = 897
Score = 54.7 bits (130), Expect = 2e-07
Identities = 40/127 (31%), Positives = 49/127 (38%), Gaps = 1/127 (0%)
Frame = +3
Query: 806 PPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASP-AAAARARHQAPQ 864
P A +PS PAA ++ PP +P A PP SP AAA A A
Sbjct: 108 PSAAPTTSPSTPAATTPVSSPVAAPPTTPTTPAPVASPKSSPPATSPKAAAPTATPPAAS 287
Query: 865 PPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPTK 924
PPA + P PP+PA + + P A + P PA PSK K
Sbjct: 288 SPPAVTPVSTPPPAPVPVKSPPTPAPVS-SPPAVTPVAAPTTTPAVPAPAPSKGKKNKKK 464
Query: 925 KPATKPS 931
A PS
Sbjct: 465 HGAPAPS 485
Score = 44.7 bits (104), Expect = 2e-04
Identities = 30/83 (36%), Positives = 34/83 (40%), Gaps = 15/83 (18%)
Frame = +3
Query: 804 PLP-PAKKQPTPSQ--------PAAAAIATNQAPQPPPSPAAAARARHQAPPP------P 848
P P P K PTP+ P AA T P P PS + +H AP P P
Sbjct: 324 PAPVPVKSPPTPAPVSSPPAVTPVAAPTTTPAVPAPAPSKGKKNKKKHGAPAPSPALLGP 503
Query: 849 PASPAAAARARHQAPQPPPASPA 871
PA PA A A P PA+ A
Sbjct: 504 PAPPAGAPGPSEDASSPGPATTA 572
Score = 43.5 bits (101), Expect = 4e-04
Identities = 31/94 (32%), Positives = 39/94 (40%), Gaps = 8/94 (8%)
Frame = +3
Query: 807 PAKKQPTPSQPAAAAI-ATNQAPQPPPSPAAAARARHQAP--PPPPASPAAAARARHQAP 863
P PT + PAA++ A PPP+P AP PP +P AA P
Sbjct: 246 PKAAAPTATPPAASSPPAVTPVSTPPPAPVPVKSPPTPAPVSSPPAVTPVAAPTTTPAVP 425
Query: 864 QPPPASPAAAARARHQAPQPP-----PPSPAAAA 892
P P S + +H AP P PP+P A A
Sbjct: 426 APAP-SKGKKNKKKHGAPAPSPALLGPPAPPAGA 524
Score = 42.4 bits (98), Expect = 8e-04
Identities = 31/122 (25%), Positives = 38/122 (30%)
Frame = +3
Query: 807 PAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQPP 866
P P S AAA AT A PP+ + PPP P + PP
Sbjct: 219 PKSSPPATSPKAAAPTATPPAASSPPAVTPVST------PPPAPVPVKSPPTPAPVSSPP 380
Query: 867 PASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPTKKP 926
+P AA P P P + P+ P PA P + P
Sbjct: 381 AVTPVAAPTTTPAVPAPAPSKGKKNKKKHGAPAPSPALLGPPAPPAGAPGPSEDASSPGP 560
Query: 927 AT 928
AT
Sbjct: 561 AT 566
Score = 41.6 bits (96), Expect = 0.001
Identities = 30/94 (31%), Positives = 36/94 (37%), Gaps = 10/94 (10%)
Frame = +3
Query: 806 PPAKKQPTPSQPAAAAIATNQAPQPPPSPAA----AARARHQAPPPPPASPAAAARARHQ 861
PPA + PA + + P P SP A AA A P P S + +H
Sbjct: 291 PPAVTPVSTPPPAPVPVKSPPTPAPVSSPPAVTPVAAPTTTPAVPAPAPSKGKKNKKKHG 470
Query: 862 APQP------PPASPAAAARARHQAPQPPPPSPA 889
AP P PPA PA A A P P + A
Sbjct: 471 APAPSPALLGPPAPPAGAPGPSEDASSPGPATTA 572
Score = 40.4 bits (93), Expect = 0.003
Identities = 36/108 (33%), Positives = 40/108 (36%), Gaps = 16/108 (14%)
Frame = +3
Query: 806 PPAKKQP---TP-SQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAA----- 856
PPA P TP S P A + P P P + A AP PA PA A
Sbjct: 273 PPAASSPPAVTPVSTPPPAPVPVKSPPTPAPVSSPPAVTPVAAPTTTPAVPAPAPSKGKK 452
Query: 857 -RARHQAPQP------PPASPAAAARARHQAPQPPPPSPAAAARARHE 897
+ +H AP P PPA PA A P SP A A E
Sbjct: 453 NKKKHGAPAPSPALLGPPAPPAGAP-----GPSEDASSPGPATTANDE 581
>TC79555 homologue to GP|10567859|gb|AAG18593.1 L8019.5 {Leishmania major},
partial (1%)
Length = 1410
Score = 54.3 bits (129), Expect = 2e-07
Identities = 33/114 (28%), Positives = 48/114 (41%)
Frame = +2
Query: 806 PPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQP 865
PP +P+P+Q + + ++ + P +P A PPPPP PA R P
Sbjct: 554 PPPTSKPSPTQTPSRSPSSARTPSTSSTPTRAPLLS-SPPPPPPFFPARRPGTRSLPKSP 730
Query: 866 PPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPA 919
P PA AR +P+ P AAAA A+R + A + +PA
Sbjct: 731 PRPRPARLPSARRASPRGGRPLAAAAAAGATTSTTASRPTRSARTSAARRGRPA 892
Score = 54.3 bits (129), Expect = 2e-07
Identities = 38/133 (28%), Positives = 50/133 (37%), Gaps = 9/133 (6%)
Frame = +2
Query: 805 LPPAKKQPT--------PSQPAAAAIATNQAPQPPPSPAAA-ARARHQAPPPPPASPAAA 855
+PP QP+ PS+P++ A T P PSP +R+ A P +S
Sbjct: 467 VPP*SPQPSSTRSSPAPPSRPSSPATTTRPPPTSKPSPTQTPSRSPSSARTPSTSSTPTR 646
Query: 856 ARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQP 915
A P PPP PA R PP P PA AR R +
Sbjct: 647 APLLSSPPPPPPFFPARRPGTRSLPKSPPRPRPARLPSARRASPRGGRPLAAAAAAGATT 826
Query: 916 SKPAAQPTKKPAT 928
S A++PT+ T
Sbjct: 827 STTASRPTRSART 865
Score = 46.6 bits (109), Expect = 4e-05
Identities = 37/137 (27%), Positives = 56/137 (40%), Gaps = 11/137 (8%)
Frame = +2
Query: 803 CPLPPAKKQ-----PTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAAR 857
CP P ++ T S PA A N P PP SP+++ PPA P +
Sbjct: 1034 CPSSPRRRSGRLPAATTSSPATNA---NSPPFPPASPSSSV---------PPAKPPTSKS 1177
Query: 858 ARHQAP------QPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKP 911
AR AP PP +SP +AA A + P +A R ++ S P
Sbjct: 1178 ARSPAPV*SRPSPPPSSSPGSAASASSRCSASSRARPTSAGSRRTRTTASSTSWRTCAAP 1357
Query: 912 AVQPSKPAAQPTKKPAT 928
P + ++ +++PA+
Sbjct: 1358 FSAPPRVSSASSRRPAS 1408
Score = 40.4 bits (93), Expect = 0.003
Identities = 36/145 (24%), Positives = 56/145 (37%), Gaps = 11/145 (7%)
Frame = +2
Query: 798 HNQRKCPLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAAR 857
H+ + P P +P QP++ T +P PP P++ A P P+ +R
Sbjct: 434 HHHQPSPKPTLVPP*SP-QPSS----TRSSPAPPSRPSSPATTTRPPPTSKPSPTQTPSR 598
Query: 858 ARHQAPQPPPASPAAAARARHQAPQPPPP-----------SPAAAARARHEGEPAARSAS 906
+ A P +S A +P PPPP P + R R P+AR AS
Sbjct: 599 SPSSARTPSTSSTPTRAPLL-SSPPPPPPFFPARRPGTRSLPKSPPRPRPARLPSARRAS 775
Query: 907 QPKKPAVQPSKPAAQPTKKPATKPS 931
+ + A T A++P+
Sbjct: 776 PRGGRPLAAAAAAGATTSTTASRPT 850
Score = 37.0 bits (84), Expect = 0.035
Identities = 40/162 (24%), Positives = 58/162 (35%), Gaps = 3/162 (1%)
Frame = +2
Query: 743 PKMSQDDILPPEIKRGPG---RPKKLRRREPDEPDKKNPTKLKRGGSSYTCGRCHQKGHN 799
P ++ PP P RP + P + ++P+ + +S T R
Sbjct: 488 PSSTRSSPAPPSRPSSPATTTRPPPTSKPSPTQTPSRSPSSARTPSTSSTPTRA------ 649
Query: 800 QRKCPLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARAR 859
PL + P P PA T P+ PP P PA +A RA
Sbjct: 650 ----PLLSSPPPPPPFFPARRP-GTRSLPKSPPRPR-------------PARLPSARRAS 775
Query: 860 HQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPA 901
+ +P A+ AA A A +P + +AAR G PA
Sbjct: 776 PRGGRPLAAAAAAGATTSTTASRPTRSARTSAAR---RGRPA 892
Score = 30.8 bits (68), Expect = 2.5
Identities = 19/53 (35%), Positives = 24/53 (44%)
Frame = -2
Query: 856 ARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQP 908
+RA H AP+ + +A P PPPP P HEG P+ R A P
Sbjct: 893 SRAFHAAPRKYVQTASAVMPWWKSLPLPPPPPPKV---GHHEGLPSWRLAVWP 744
>TC93060 weakly similar to PIR|S55970|S55970 ribosomal protein L6.e.B
cytosolic - yeast (Saccharomyces cerevisiae), partial
(77%)
Length = 903
Score = 54.3 bits (129), Expect = 2e-07
Identities = 48/161 (29%), Positives = 72/161 (43%), Gaps = 14/161 (8%)
Frame = +3
Query: 785 GSSYTCGRCHQKGHNQRKCPLPPAKKQPTPSQPAAAAIATNQAPQ----PPPSPAAAA-- 838
GSS R + G P P ++ PS P A ++ AP P S A+A+
Sbjct: 42 GSSLPPPRRPRSGTTPTMSPRP--RRSARPSAPGTLASPSSPAPS*FSLPVASAASASSS 215
Query: 839 -----RARHQAPPPP--PASPAAAARARHQAPQPPPASPAAAARARHQAPQPPPPSPAAA 891
R +P P ASP+A + +P P ++ + R R + + P SP
Sbjct: 216 *RPSTRVSSSSPAPSRSTASPSAVSTPATSSPHPTRSTSPLSTRRRLRRSRNPSTSPPTR 395
Query: 892 ARARHEGEPAAR-SASQPKKPAVQPSKPAAQPTKKPATKPS 931
R R PA R S+S+ + P + S AA PT++P+T+PS
Sbjct: 396 PRPR----PARRLSSSRERSPRRRRSTAAALPTRRPSTRPS 506
Score = 34.7 bits (78), Expect = 0.17
Identities = 30/102 (29%), Positives = 43/102 (41%), Gaps = 5/102 (4%)
Frame = +3
Query: 832 PSPAAAARARHQAPPPP-----PASPAAAARARHQAPQPPPASPAAAARARHQAPQPPPP 886
P P ++AR + PPP +P + R R A P++P A AP
Sbjct: 12 PRPRSSARLLGSSLPPPRRPRSGTTPTMSPRPRRSA---RPSAPGTLASPSSPAPS*FSL 182
Query: 887 SPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPTKKPAT 928
A+AA A P+ R +S P+ + P+A T PAT
Sbjct: 183 PVASAASASSS*RPSTRVSSSSPAPSRSTASPSAVST--PAT 302
>BF521488 weakly similar to GP|18447433|gb RE28280p {Drosophila
melanogaster}, partial (84%)
Length = 470
Score = 53.9 bits (128), Expect = 3e-07
Identities = 38/105 (36%), Positives = 44/105 (41%), Gaps = 3/105 (2%)
Frame = -3
Query: 829 QPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQPPPASPAAAARARHQAPQPPPPSP 888
+PPP AAA A + P PP PAA A A +A PPP P R P PPPP+P
Sbjct: 420 KPPPLAEAAADAAAEPPRKPP--PAALALAEAEAEPPPPPPPPRKGR----PPNPPPPNP 259
Query: 889 AAAARARHEGEPAARSASQPK---KPAVQPSKPAAQPTKKPATKP 930
P R P P +P +P P K P KP
Sbjct: 258 -----------P*GRPPPNPP*GL*PPPKPGRPKPPPPKPPPPKP 157
Score = 52.0 bits (123), Expect = 1e-06
Identities = 35/117 (29%), Positives = 46/117 (38%)
Frame = -3
Query: 810 KQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQPPPAS 869
++P P AAA A +PPP+ A A A + PPPPP + + P PPP +
Sbjct: 423 RKPPPLAEAAADAAAEPPRKPPPAALALAEAEAEPPPPPPPP------RKGRPPNPPPPN 262
Query: 870 PAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSKPAAQPTKKP 926
P +PPP P G P PK P +P +P P P
Sbjct: 261 PP*G--------RPPPNPP*GL*PPPKPGRP---KPPPPKPPPPKPGRPKPPPKPGP 124
Score = 45.4 bits (106), Expect = 1e-04
Identities = 30/95 (31%), Positives = 38/95 (39%), Gaps = 2/95 (2%)
Frame = -3
Query: 808 AKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQPPP 867
A + P PAA A+A +A PPP P + + P PPP +P P PP
Sbjct: 384 AAEPPRKPPPAALALAEAEAEPPPPPPPP---RKGRPPNPPPPNPP*G----RPPPNPP* 226
Query: 868 A--SPAAAARARHQAPQPPPPSPAAAARARHEGEP 900
P R + P+PPPP P G P
Sbjct: 225 GL*PPPKPGRPKPPPPKPPPPKPGRPKPPPKPGPP 121
Score = 30.4 bits (67), Expect = 3.3
Identities = 26/97 (26%), Positives = 31/97 (31%), Gaps = 6/97 (6%)
Frame = -3
Query: 804 PLPPAKKQPTPSQPAAAAIATNQAPQPP------PSPAAAARARHQAPPPPPASPAAAAR 857
P PP +K P+ P P PP P P + PPP P
Sbjct: 312 PPPPPRKGRPPNPPPPNPP*GRPPPNPP*GL*PPPKPGRPKPPPPKPPPPKPG------- 154
Query: 858 ARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARA 894
P+PPP P PP P+ AA A
Sbjct: 153 ----RPKPPP------------KPGPPKPNCGLAATA 91
>TC83704 similar to PIR|T05005|T05005 hypothetical protein T19P19.70 -
Arabidopsis thaliana, partial (15%)
Length = 716
Score = 53.5 bits (127), Expect = 4e-07
Identities = 35/101 (34%), Positives = 37/101 (35%)
Frame = +1
Query: 811 QPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAARARHQAPQPPPASP 870
+P P Q P PPP R Q PPPPP S A H P PPP S
Sbjct: 298 EPLPYQHKEQPPIPVTLPPPPPLSKLPPATREQLPPPPPLSKLPPAAREH--PPPPPPSA 471
Query: 871 AAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKP 911
AR + P PPP S PAAR P P
Sbjct: 472 KLPPAARERLPLPPPLSKL---------PPAARERLPPPSP 567
Score = 51.2 bits (121), Expect = 2e-06
Identities = 33/96 (34%), Positives = 40/96 (41%)
Frame = +1
Query: 798 HNQRKCPLPPAKKQPTPSQPAAAAIATNQAPQPPPSPAAAARARHQAPPPPPASPAAAAR 857
+ ++ P P P P Q P PPP AR PPPPP++ A
Sbjct: 310 YQHKEQPPIPVTLPPPPPLSKLPPATREQLPPPPPLSKLPPAAREHPPPPPPSAKLPPA- 486
Query: 858 ARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAAR 893
AR + P PPP S A AR + P PP P P R
Sbjct: 487 ARERLPLPPPLSKLPPA-ARERLP-PPSPLPEKVDR 588
Score = 50.1 bits (118), Expect = 4e-06
Identities = 36/129 (27%), Positives = 43/129 (32%), Gaps = 8/129 (6%)
Frame = +1
Query: 806 PPAKKQPTPSQPAAAAIATNQAPQP--------PPSPAAAARARHQAPPPPPASPAAAAR 857
PP + +AT P+P PP P PPPPP S A
Sbjct: 229 PPPPVAFANNDSTVPPVATTSLPEPLPYQHKEQPPIPVTL-------PPPPPLSKLPPA- 384
Query: 858 ARHQAPQPPPASPAAAARARHQAPQPPPPSPAAAARARHEGEPAARSASQPKKPAVQPSK 917
R Q P PPP S A H P PP AAR R P + + P
Sbjct: 385 TREQLPPPPPLSKLPPAAREHPPPPPPSAKLPPAARERLPLPPPLSKLPPAARERLPPPS 564
Query: 918 PAAQPTKKP 926
P + +P
Sbjct: 565 PLPEKVDRP 591
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.138 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 32,470,791
Number of Sequences: 36976
Number of extensions: 633191
Number of successful extensions: 17461
Number of sequences better than 10.0: 701
Number of HSP's better than 10.0 without gapping: 5455
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 9861
length of query: 931
length of database: 9,014,727
effective HSP length: 105
effective length of query: 826
effective length of database: 5,132,247
effective search space: 4239236022
effective search space used: 4239236022
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0016.18


