

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0013.6
(937 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
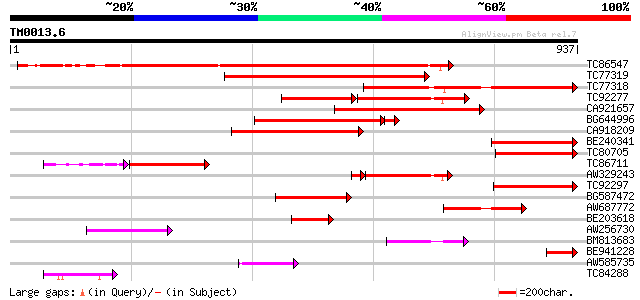
Score E
Sequences producing significant alignments: (bits) Value
TC86547 similar to GP|20805911|gb|AAM28891.1 NADPH oxidase {Nico... 561 e-160
TC77319 similar to GP|16549089|dbj|BAB70751. respiratory burst o... 427 e-120
TC77318 similar to GP|16549089|dbj|BAB70751. respiratory burst o... 414 e-116
TC92277 similar to GP|16549089|dbj|BAB70751. respiratory burst o... 227 1e-95
CA921657 similar to PIR|T03826|T03 cytochrome b245 beta chain ho... 315 5e-86
BG644996 similar to GP|20805911|gb NADPH oxidase {Nicotiana taba... 249 1e-71
CA918209 weakly similar to GP|16549089|db respiratory burst oxid... 256 2e-68
BE240341 similar to GP|16549089|db respiratory burst oxidase hom... 206 3e-53
TC80705 similar to GP|19715905|emb|CAC84140. NADPH oxidase {Nico... 205 6e-53
TC86711 similar to GP|16549089|dbj|BAB70751. respiratory burst o... 179 3e-49
AW329243 similar to GP|16549089|db respiratory burst oxidase hom... 169 4e-46
TC92297 weakly similar to GP|19715905|emb|CAC84140. NADPH oxidas... 182 5e-46
BG587472 similar to GP|20160800|db putative respiratory burst ox... 157 2e-38
AW687772 similar to GP|16549089|db respiratory burst oxidase hom... 124 2e-28
BE203618 weakly similar to GP|3242787|gb|A respiratory burst oxi... 119 7e-27
AW256730 weakly similar to GP|9757922|dbj respiratory burst oxid... 94 2e-19
BM813683 weakly similar to GP|15341529|gb ferric-chelate reducta... 74 2e-13
BE941228 similar to GP|20160800|dbj putative respiratory burst o... 60 4e-09
AW585735 similar to GP|13958313|gb| NADPH oxidase 1 {Podospora a... 58 2e-08
TC84288 similar to GP|17384016|emb|CAC87256. NADPH oxidase {Nico... 52 8e-07
>TC86547 similar to GP|20805911|gb|AAM28891.1 NADPH oxidase {Nicotiana
tabacum}, partial (57%)
Length = 2214
Score = 561 bits (1446), Expect = e-160
Identities = 332/737 (45%), Positives = 445/737 (60%), Gaps = 16/737 (2%)
Frame = +1
Query: 13 DIETDSPENLRTSSPPASEGHAPSHGYSSMLPVFLNDLRTNHHKELVEITLELEDDAVVL 72
DIE +PEN S S+ S+ V D + VE+T++++ D+V L
Sbjct: 109 DIELIAPENSTKLSTTESK---------SVTDV---DHNIEEDADYVEVTMDIQGDSVAL 252
Query: 73 CNVAPASS---GAGTSSSSSHATGDEGTSASGAGAVARSLSKNLSITSRIRRKFPWLRSM 129
+V + G G S TG++ GA S+ +N +I ++ +
Sbjct: 253 HSVKTVTESDMGEGEHEKQS-LTGNKLEKKKSFGA---SVVQNATIR---------IKQL 393
Query: 130 SMRTSASSTESVEDPVESARNARRMQAKLERTRSSAQRALKGLRFISKSGEAGEELWRKV 189
S S E + +LERT+S+ AL GL+FISK+ W +V
Sbjct: 394 KRLASFSKPEPAK--------------RLERTKSAVAHALTGLKFISKTDVGAG--WSEV 525
Query: 190 EERFGSLA--KDGLLAREDFGGCIGM-DDSKEFAVGVFDALARRKGRKASKITKEELHHF 246
E+ F L DG L R F CIG+ ++SK +A +FD LAR++G + I K + F
Sbjct: 526 EKVFDKLTVTTDGYLPRTLFAKCIGLNEESKAYAEMLFDTLARQRGIQGGSINKIQFREF 705
Query: 247 WQQISDQSFDARLQIFFDMADSNEDGRITREEVQELIMLSASANKLSKLKEQAEGYAALI 306
W ISDQSFD RL+IFFDM D + DGRIT EE++ +I LSA+ANKLS +++QAE YAALI
Sbjct: 706 WDCISDQSFDTRLKIFFDMVDKDADGRITEEEIKNIICLSATANKLSNIQKQAEEYAALI 885
Query: 307 MEELDPENLGYIELWQLEMLLLQ--KDRYMNYSRQLSTSSVSWSQNMTGLKHKNEIQRLC 364
MEELDP++ GYI + LE LLL ++ S+ LS SQ + N I+R
Sbjct: 886 MEELDPDDTGYILIGNLETLLLHGPEETTRGESKYLSQML---SQKLRPTFEGNPIKRWY 1056
Query: 365 RTLQCLALEYWRRGWVLFLWLVTTASLFAWKFYQYRNRTSFQVMGYCLPVAKGAAETLKL 424
R + + WRR WV LW+ LFA+KF QYR R++++VMG+C+ +AKGAAETLKL
Sbjct: 1057RDTKYFFQDNWRRSWVFALWIGVMLGLFAFKFVQYRRRSAYKVMGHCVCMAKGAAETLKL 1236
Query: 425 NMALILLPVCRNTLTWLRS-TRARKIVPFDDNINFHKIIACAIVVGVVVHAGNHLACDFP 483
NMA+ILLPVCRNT+TWLR+ T+ VPFDD +NFHK+IA AI +GV +HA HLA DFP
Sbjct: 1237NMAIILLPVCRNTITWLRNKTKLGAFVPFDDGLNFHKMIALAIAIGVGIHAIYHLA*DFP 1416
Query: 484 LLINSSPEEFSLIASDFNNKKPTYKTLLTGVEGVTGISMVTLMAISFTLATHHFRRNAVR 543
++++S E++ LI F K Y + EGVTGI MV LMAI+FTLA FRRN +
Sbjct: 1417RILHASNEKYKLIEPFFGEKPTNYWHFVKSWEGVTGILMVVLMAIAFTLANTRFRRNRTK 1596
Query: 544 LPPPLNRLTGFNAFWYSHHLLGVVYILLFIHGSFLNLTHRWYNKTTWMYISVPLLLYIAE 603
LP P N+LTGFNAFWYSHHL +VY +L IHG+ L LT W +KTTWMY+++P+ +Y E
Sbjct: 1597LPKPFNKLTGFNAFWYSHHLFIIVYAMLIIHGTKLYLTKEWNHKTTWMYLAIPITIYGLE 1776
Query: 604 RTLRTSRSKHYAVKIQKVSVLPGNVFSLIMSKPNGFKYKSGQYIFLQCPKISPFEWHPFS 663
R +R RS +V+I KV+V PGNV ++ MSKP GF YKSGQY+ + C +SP EWHPFS
Sbjct: 1777RLIRALRSSIKSVRILKVAVYPGNVLAINMSKPQGFSYKSGQYMLVNCAAVSPLEWHPFS 1956
Query: 664 ITSAPGDEYLSVHIRTVGDWTQELKLLFTEDDIPPVN--CNATFGELVQ----MDQREHP 717
ITSAP D+YLSVHI+ +GDWT+ LK F +P + TF EL Q P
Sbjct: 1957ITSAPNDDYLSVHIKILGDWTRSLKTKFL-TGLPASHQWTEWTF*ELNA*KEITAQVLFP 2133
Query: 718 RLFVDGP-YGAPAQDFK 733
++ +DGP + PAQD++
Sbjct: 2134KVLIDGPVWERPAQDYR 2184
>TC77319 similar to GP|16549089|dbj|BAB70751. respiratory burst oxidase
homolog {Solanum tuberosum}, partial (42%)
Length = 1137
Score = 427 bits (1098), Expect = e-120
Identities = 201/341 (58%), Positives = 257/341 (74%), Gaps = 2/341 (0%)
Frame = +2
Query: 355 KHKNEIQRLCRTLQCLALEYWRRGWVLFLWLVTTASLFAWKFYQYRNRTSFQVMGYCLPV 414
K N I+R R+L + W+R WV+ LWL A+LF WKF QY+NR++F+VMGYC+ +
Sbjct: 8 KEYNPIKRSLRSLNYFIEDNWKRIWVIALWLSICAALFTWKFIQYKNRSAFRVMGYCVTI 187
Query: 415 AKGAAETLKLNMALILLPVCRNTLTWLRS-TRARKIVPFDDNINFHKIIACAIVVGVVVH 473
AKGAAETLK NMALIL+PVCRNT+TWLRS T+ +VPFDDNINFHK+IA I +GV +H
Sbjct: 188 AKGAAETLKFNMALILMPVCRNTITWLRSKTKLGVVVPFDDNINFHKVIAFGIAIGVGLH 367
Query: 474 AGNHLACDFPLLINSSPEEFSLIASDFNNKKPT-YKTLLTGVEGVTGISMVTLMAISFTL 532
A +HL CDFP L++++ E+ + F + +P Y + G EG TGI MV LMAI+FTL
Sbjct: 368 AISHLTCDFPRLLHATDAEYVPMKKFFGDHRPNNYWWFVKGTEGWTGIVMVVLMAIAFTL 547
Query: 533 ATHHFRRNAVRLPPPLNRLTGFNAFWYSHHLLGVVYILLFIHGSFLNLTHRWYNKTTWMY 592
A FRRN ++LPP L +LTGFNAFWYSHHL +VY LL +HG FL L+ +WY KTTWMY
Sbjct: 548 AQPWFRRNKLKLPPLLKKLTGFNAFWYSHHLFVIVYALLIVHGYFLYLSKKWYKKTTWMY 727
Query: 593 ISVPLLLYIAERTLRTSRSKHYAVKIQKVSVLPGNVFSLIMSKPNGFKYKSGQYIFLQCP 652
+++P+++Y ER LR RS + +VKI KV+V PGNV +L +SKP GFKY SGQYIF+ C
Sbjct: 728 LAIPMIIYACERLLRAFRSGYKSVKILKVAVYPGNVLALHVSKPQGFKYHSGQYIFVNCA 907
Query: 653 KISPFEWHPFSITSAPGDEYLSVHIRTVGDWTQELKLLFTE 693
+SPF+WHPFSITSAPGD+Y+SVHIRT GDWT +LK +F +
Sbjct: 908 DVSPFQWHPFSITSAPGDDYVSVHIRTAGDWTSQLKAVFAK 1030
>TC77318 similar to GP|16549089|dbj|BAB70751. respiratory burst oxidase
homolog {Solanum tuberosum}, partial (38%)
Length = 1394
Score = 414 bits (1064), Expect = e-116
Identities = 209/362 (57%), Positives = 259/362 (70%), Gaps = 10/362 (2%)
Frame = +2
Query: 586 NKTTWMYISVPLLLYIAERTLRTSRSKHYAVKIQKVSVLPGNVFSLIMSKPNGFKYKSGQ 645
NKTTWMY+++P+++Y ER LR RS + +VKI KV+V PGNV +L +SKP GFKY SGQ
Sbjct: 77 NKTTWMYLAIPMIIYACERLLRAFRSGYKSVKILKVAVYPGNVLALHVSKPQGFKYHSGQ 256
Query: 646 YIFLQCPKISPFEWHPFSITSAPGDEYLSVHIRTVGDWTQELKLLFTEDDIPPVNCNATF 705
YIF+ C +SPF+WHPFSITSAPGD+Y+SVHIRT GDWT +LK +F + C
Sbjct: 257 YIFVNCADVSPFQWHPFSITSAPGDDYVSVHIRTAGDWTSQLKAVFAKA------CQPAS 418
Query: 706 GE---LVQMDQREH------PRLFVDGPYGAPAQDFKNFDVLLLIGLGIGATPFISILRD 756
G+ L++ D + P+L +DGPYGAPAQD+K+++V+LL+GLGIGATP ISIL+D
Sbjct: 419 GDQSGLLRADMLQGNNIPRMPKLLIDGPYGAPAQDYKDYEVILLVGLGIGATPLISILKD 598
Query: 757 LLNNTRASDESMDQSNTETTRSDESWNSFTSSNLTPGGNKRSQRTTN-AYFYWVTREPGS 815
+LNN + + ++Q E+ + NKR TN AYFYWVTRE GS
Sbjct: 599 VLNNMK-QQKDIEQGVVESGVKN---------------NKRKPFATNRAYFYWVTREQGS 730
Query: 816 FEWFKGVMDEVAEMDHRGQIELHNYLTSVYEEGDARSTLITMIQALNHAKHGVDILSGTR 875
FEWFKGVMDE+A+ D G IELHNY TSVYEEGDARS LITM+Q+L+HAK GVDI+SGTR
Sbjct: 731 FEWFKGVMDEIADYDKDGLIELHNYCTSVYEEGDARSALITMLQSLHHAKSGVDIVSGTR 910
Query: 876 VRTHFARPNWREVFIKIASKHPYSTVGVFYCGMPVLAKELKKLSLELSHKTTTRFDFHKE 935
V+THFARPNWR VF A KHP VGVFYCG L +LK LSL+ S KT T+F+FHKE
Sbjct: 911 VKTHFARPNWRTVFKHTALKHPGKRVGVFYCGAAGLVGQLKSLSLDFSRKTNTKFEFHKE 1090
Query: 936 YF 937
F
Sbjct: 1091NF 1096
Score = 40.0 bits (92), Expect = 0.004
Identities = 18/36 (50%), Positives = 24/36 (66%), Gaps = 1/36 (2%)
Frame = -2
Query: 561 HHLLGVVYILLFI-HGSFLNLTHRWYNKTTWMYISV 595
+H G ++ FI HG FL L+ +WY KTTWMYI +
Sbjct: 115 YHRNGKIHPSRFIVHGYFLYLSKKWYKKTTWMYIII 8
>TC92277 similar to GP|16549089|dbj|BAB70751. respiratory burst oxidase
homolog {Solanum tuberosum}, partial (34%)
Length = 949
Score = 227 bits (579), Expect(2) = 1e-95
Identities = 112/191 (58%), Positives = 141/191 (73%), Gaps = 7/191 (3%)
Frame = +3
Query: 576 SFLNLTHRWYNKTTWMYISVPLLLYIAERTLRTSRSKHYAVKIQKVSVLPGNVFSLIMSK 635
+FL L+ +WY KTTWMY++VP++LY ER L RS + +V+I KV+V PGNV +L SK
Sbjct: 384 TFLYLSKKWYKKTTWMYLAVPMILYGCERLLPAFRSGYKSVRILKVAVYPGNVLALHASK 563
Query: 636 PNGFKYKSGQYIFLQCPKISPFEWHPFSITSAPGDEYLSVHIRTVGDWTQELKLLFTEDD 695
P GFKY SGQYI++ C ISPFEWHPFSITSAPGD+Y+SVHIRT+GDWT +LK +F +
Sbjct: 564 PQGFKYTSGQYIYVNCSDISPFEWHPFSITSAPGDDYISVHIRTLGDWTSQLKGIFAKAC 743
Query: 696 IPPVNCNATFGELVQMDQ-------REHPRLFVDGPYGAPAQDFKNFDVLLLIGLGIGAT 748
P N L++ D PRL +DGPYGAPAQD+KN++VLLL+GLGIGAT
Sbjct: 744 QP---ANDDQSGLLRADMLPGKSSLPRMPRLRIDGPYGAPAQDYKNYEVLLLVGLGIGAT 914
Query: 749 PFISILRDLLN 759
P ISIL+D+LN
Sbjct: 915 PLISILKDVLN 947
Score = 142 bits (358), Expect(2) = 1e-95
Identities = 67/124 (54%), Positives = 89/124 (71%), Gaps = 1/124 (0%)
Frame = +1
Query: 450 VPFDDNINFHKIIACAIVVGVVVHAGNHLACDFPLLINSSPEEFSLIASDFNNKKPT-YK 508
VPFDDNINFHK+IA I +GV +HA +HL CDFP L++++ EE+ + F +++P Y
Sbjct: 1 VPFDDNINFHKVIAFGIAIGVGLHAISHLTCDFPRLLHATDEEYEPMKQFFGDERPNNYW 180
Query: 509 TLLTGVEGVTGISMVTLMAISFTLATHHFRRNAVRLPPPLNRLTGFNAFWYSHHLLGVVY 568
+ G EG TG+ MV LMAI+F LA FRRN + LP PL +LTGFNAFWYSHHL +VY
Sbjct: 181 WFVKGTEGWTGVVMVVLMAIAFILAQPWFRRNRLNLPKPLKKLTGFNAFWYSHHLFVIVY 360
Query: 569 ILLF 572
+ ++
Sbjct: 361 VPIY 372
>CA921657 similar to PIR|T03826|T03 cytochrome b245 beta chain homolog
RbohAp108 - Arabidopsis thaliana, partial (26%)
Length = 749
Score = 315 bits (807), Expect = 5e-86
Identities = 150/247 (60%), Positives = 188/247 (75%)
Frame = -2
Query: 538 RRNAVRLPPPLNRLTGFNAFWYSHHLLGVVYILLFIHGSFLNLTHRWYNKTTWMYISVPL 597
RRN ++LP P +RLTGFNAFWYSHHL +VY+LL IHG L L H+WY +TTWMY++VPL
Sbjct: 748 RRNLIKLPEPFSRLTGFNAFWYSHHLFVIVYVLLIIHGVKLYLVHKWYYQTTWMYLTVPL 569
Query: 598 LLYIAERTLRTSRSKHYAVKIQKVSVLPGNVFSLIMSKPNGFKYKSGQYIFLQCPKISPF 657
LLY +ERTLR RS Y V++ KV++ PGNV +L MSKP F+YKSGQY+F+QC +SPF
Sbjct: 568 LLYASERTLRLFRSGLYTVRLIKVAIYPGNVLTLQMSKPPQFRYKSGQYMFVQCAAVSPF 389
Query: 658 EWHPFSITSAPGDEYLSVHIRTVGDWTQELKLLFTEDDIPPVNCNATFGELVQMDQREHP 717
EWHPFSITSAPGD+YLSVHIR +GDWTQELK +F+E PPV+ + + ++ P
Sbjct: 388 EWHPFSITSAPGDDYLSVHIRQLGDWTQELKRVFSEVCEPPVSGRSGLLRADETTKKSLP 209
Query: 718 RLFVDGPYGAPAQDFKNFDVLLLIGLGIGATPFISILRDLLNNTRASDESMDQSNTETTR 777
+L +DGPYGAPAQD++ +DVLLL+GLGIGATPFISIL+DLLNN +E D + +
Sbjct: 208 KLKIDGPYGAPAQDYRKYDVLLLVGLGIGATPFISILKDLLNNIVKMEELADSVSDTSRG 29
Query: 778 SDESWNS 784
SD S S
Sbjct: 28 SDLSVGS 8
>BG644996 similar to GP|20805911|gb NADPH oxidase {Nicotiana tabacum},
partial (25%)
Length = 726
Score = 249 bits (637), Expect(2) = 1e-71
Identities = 121/217 (55%), Positives = 156/217 (71%), Gaps = 1/217 (0%)
Frame = +3
Query: 405 FQVMGYCLPVAKGAAETLKLNMALILLPVCRNTLTWLRS-TRARKIVPFDDNINFHKIIA 463
++VMG+C+ +AKGAAETLKLNMA+ILLPVCRNT+TWLR+ T+ VPFDDN+NFHK+IA
Sbjct: 3 YEVMGHCVCMAKGAAETLKLNMAIILLPVCRNTITWLRNKTKLGIAVPFDDNLNFHKVIA 182
Query: 464 CAIVVGVVVHAGNHLACDFPLLINSSPEEFSLIASDFNNKKPTYKTLLTGVEGVTGISMV 523
A+ GV +HA HL CDFP L++++ E++ L+ F + +Y + EGVTGI MV
Sbjct: 183 VAVATGVGIHAIYHLTCDFPRLLHANSEKYKLMEPFFGKQPTSYWHFVKSWEGVTGIVMV 362
Query: 524 TLMAISFTLATHHFRRNAVRLPPPLNRLTGFNAFWYSHHLLGVVYILLFIHGSFLNLTHR 583
LM I+FTLA+ FR+ V+LP PLN LTGFNAFWYSHHL VY LL +HG L LT
Sbjct: 363 VLMTIAFTLASPWFRKGRVKLPKPLNSLTGFNAFWYSHHLFVFVYALLVVHGIKLYLTRE 542
Query: 584 WYNKTTWMYISVPLLLYIAERTLRTSRSKHYAVKIQK 620
W+ KTTWMY+ +P+++Y ER R RS V+I K
Sbjct: 543 WHKKTTWMYLVIPIIIYALERLTRALRSSIKPVRILK 653
Score = 40.0 bits (92), Expect(2) = 1e-71
Identities = 17/25 (68%), Positives = 21/25 (84%)
Frame = +1
Query: 620 KVSVLPGNVFSLIMSKPNGFKYKSG 644
+V+V PGNV +L MSKP GF+YKSG
Sbjct: 652 RVAVYPGNVLALHMSKPQGFRYKSG 726
>CA918209 weakly similar to GP|16549089|db respiratory burst oxidase homolog
{Solanum tuberosum}, partial (20%)
Length = 713
Score = 256 bits (655), Expect = 2e-68
Identities = 124/220 (56%), Positives = 154/220 (69%), Gaps = 2/220 (0%)
Frame = +3
Query: 367 LQCLALEYWRRGWVLFLWLVTTASLFAWKFYQYRNRTSFQVMGYCLPVAKGAAETLKLNM 426
++ L YWRR WV+ LWLV LF WKF QY R+ F+VMGYCLP AKGAAETLKLNM
Sbjct: 48 VEVLFRTYWRRSWVVVLWLVGCFGLFGWKFDQYSKRSGFEVMGYCLPTAKGAAETLKLNM 227
Query: 427 ALILLPVCRNTLTWLRS-TRARKIVPFDDNINFHKIIACAIVVGVVVHAGNHLACDFPLL 485
ALILLPVCRNT+TWLR+ R ++PF+DNINFHK+IA IVVGV++H G HL CDFP +
Sbjct: 228 ALILLPVCRNTITWLRNDPRLNYVIPFNDNINFHKLIAGGIVVGVILHGGTHLTCDFPRI 407
Query: 486 INSSPEEF-SLIASDFNNKKPTYKTLLTGVEGVTGISMVTLMAISFTLATHHFRRNAVRL 544
+S F IA+ F +PTY +L E +GI MV +MAI+F+LAT RR + L
Sbjct: 408 SDSDKSIFRQTIAAGFGYHQPTYMEILATTEVASGIGMVLIMAIAFSLATKWPRRRSPVL 587
Query: 545 PPPLNRLTGFNAFWYSHHLLGVVYILLFIHGSFLNLTHRW 584
P + R+TG+N FWYSHHL +VY+LL +H FL L +W
Sbjct: 588 PLSIRRVTGYNTFWYSHHLFVIVYVLLIVHSMFLFLADKW 707
>BE240341 similar to GP|16549089|db respiratory burst oxidase homolog
{Solanum tuberosum}, partial (16%)
Length = 569
Score = 206 bits (524), Expect = 3e-53
Identities = 99/142 (69%), Positives = 112/142 (78%)
Frame = +1
Query: 796 KRSQRTTNAYFYWVTREPGSFEWFKGVMDEVAEMDHRGQIELHNYLTSVYEEGDARSTLI 855
KR T AYFYWVTRE GSFEW KGVM+EVAE D G IELHNY TSVYEEGDARS LI
Sbjct: 10 KRPFATKRAYFYWVTREQGSFEWLKGVMNEVAENDKEGVIELHNYCTSVYEEGDARSALI 189
Query: 856 TMIQALNHAKHGVDILSGTRVRTHFARPNWREVFIKIASKHPYSTVGVFYCGMPVLAKEL 915
TM+Q+L+HAK+GVD++S TRV+THFARPNWR V+ +A KHP VGVFYCG L EL
Sbjct: 190 TMLQSLHHAKNGVDVVSETRVKTHFARPNWRNVYKHVALKHPEKKVGVFYCGAHGLVGEL 369
Query: 916 KKLSLELSHKTTTRFDFHKEYF 937
+KLSL+ S KT T+FDFHKE F
Sbjct: 370 RKLSLDFSRKTGTKFDFHKENF 435
>TC80705 similar to GP|19715905|emb|CAC84140. NADPH oxidase {Nicotiana
tabacum}, partial (14%)
Length = 837
Score = 205 bits (522), Expect = 6e-53
Identities = 92/134 (68%), Positives = 112/134 (82%)
Frame = +1
Query: 804 AYFYWVTREPGSFEWFKGVMDEVAEMDHRGQIELHNYLTSVYEEGDARSTLITMIQALNH 863
AYFYWVTRE GSF+WFKGVM++VAE D RG IELH+Y TSVYE+GDARS LI M+Q++NH
Sbjct: 1 AYFYWVTREQGSFDWFKGVMNDVAEEDRRGLIELHSYCTSVYEQGDARSALIAMVQSINH 180
Query: 864 AKHGVDILSGTRVRTHFARPNWREVFIKIASKHPYSTVGVFYCGMPVLAKELKKLSLELS 923
AKHGVD++SGTRV +HFA+PNWR V+ +IA HP + VGVFYCG L EL++LSL+ S
Sbjct: 181 AKHGVDVVSGTRVMSHFAKPNWRTVYKRIALNHPEAQVGVFYCGPSTLTHELRQLSLDFS 360
Query: 924 HKTTTRFDFHKEYF 937
H T+T+FDFHKE F
Sbjct: 361 HNTSTKFDFHKENF 402
>TC86711 similar to GP|16549089|dbj|BAB70751. respiratory burst oxidase
homolog {Solanum tuberosum}, partial (27%)
Length = 1075
Score = 179 bits (453), Expect(2) = 3e-49
Identities = 89/131 (67%), Positives = 109/131 (82%)
Frame = +2
Query: 199 DGLLAREDFGGCIGMDDSKEFAVGVFDALARRKGRKASKITKEELHHFWQQISDQSFDAR 258
DG L + F CIGM++SK+FA +FDALARR+G ++ ITK+EL FW+QI+DQSFD+R
Sbjct: 656 DGKLPKTRFSQCIGMNESKDFAGELFDALARRRGITSASITKDELRQFWEQITDQSFDSR 835
Query: 259 LQIFFDMADSNEDGRITREEVQELIMLSASANKLSKLKEQAEGYAALIMEELDPENLGYI 318
LQ FFDM D N DGRI+ +EV+E+I LSASANKLSKL+E+AE YAALIMEELDP+NLG+I
Sbjct: 836 LQTFFDMVDKNADGRISEDEVKEIITLSASANKLSKLQERAEEYAALIMEELDPDNLGFI 1015
Query: 319 ELWQLEMLLLQ 329
EL LEMLLLQ
Sbjct: 1016ELHNLEMLLLQ 1048
Score = 35.8 bits (81), Expect(2) = 3e-49
Identities = 33/141 (23%), Positives = 63/141 (44%)
Frame = +1
Query: 56 KELVEITLELEDDAVVLCNVAPASSGAGTSSSSSHATGDEGTSASGAGAVARSLSKNLSI 115
++ VEITL++ DD V + N+ GD T+ +
Sbjct: 346 QDYVEITLDVRDDTVSVQNIRG---------------GDSETAL---------------L 435
Query: 116 TSRIRRKFPWLRSMSMRTSASSTESVEDPVESARNARRMQAKLERTRSSAQRALKGLRFI 175
SR+ ++ T + + V ++ ++++ +++R +S A RALKGL+F+
Sbjct: 436 ASRLEKR--------PSTLSVKLKQVSQELKRMTSSKKFD-RVDRAKSGAARALKGLKFM 588
Query: 176 SKSGEAGEELWRKVEERFGSL 196
+K+ + W +VE+RF L
Sbjct: 589 TKN-VGTDRGWSQVEKRFDEL 648
>AW329243 similar to GP|16549089|db respiratory burst oxidase homolog
{Solanum tuberosum}, partial (18%)
Length = 627
Score = 169 bits (428), Expect(2) = 4e-46
Identities = 85/150 (56%), Positives = 105/150 (69%), Gaps = 7/150 (4%)
Frame = +3
Query: 589 TWMYISVPLLLYIAERTLRTSRSKHYAVKIQKVSVLPGNVFSLIMSKPNGFKYKSGQYIF 648
TWMY++VP++LY ER LR RS + +V+I KV+V PGNV +L SKP GFKY SGQYI+
Sbjct: 186 TWMYLAVPMILYGCERLLRAFRSGYKSVRILKVAVYPGNVLALHASKPQGFKYTSGQYIY 365
Query: 649 LQCPKISPFEWHPFSITSAPGDEYLSVHIRTVGDWTQELKLLFTEDDIPPVNCNATFGEL 708
+ C ISPFEWHPFSITSAPGD+Y+SVHIRT+GDWT +LK +F + P N L
Sbjct: 366 VNCSDISPFEWHPFSITSAPGDDYISVHIRTLGDWTSQLKGIFAKACQP---ANDDQSGL 536
Query: 709 VQMDQ-------REHPRLFVDGPYGAPAQD 731
++ D PRL +DGPYGAPAQD
Sbjct: 537 LRADMLPGKSSLPRMPRLRIDGPYGAPAQD 626
Score = 35.0 bits (79), Expect(2) = 4e-46
Identities = 13/23 (56%), Positives = 17/23 (73%)
Frame = +2
Query: 566 VVYILLFIHGSFLNLTHRWYNKT 588
+VY+L IHG FL L+ +WY KT
Sbjct: 8 IVYVLFIIHGYFLYLSKKWYKKT 76
>TC92297 weakly similar to GP|19715905|emb|CAC84140. NADPH oxidase
{Nicotiana tabacum}, partial (14%)
Length = 719
Score = 182 bits (462), Expect = 5e-46
Identities = 80/138 (57%), Positives = 107/138 (76%)
Frame = +3
Query: 800 RTTNAYFYWVTREPGSFEWFKGVMDEVAEMDHRGQIELHNYLTSVYEEGDARSTLITMIQ 859
+T AYFYW+ E G F+WFKGV++EVAE DH+ IE+H++LTSVY +GDARS L+T++Q
Sbjct: 30 KTRKAYFYWMAGEQGFFDWFKGVINEVAEEDHKKVIEIHSHLTSVYXDGDARSALVTVLQ 209
Query: 860 ALNHAKHGVDILSGTRVRTHFARPNWREVFIKIASKHPYSTVGVFYCGMPVLAKELKKLS 919
+LNHAK G+DIL+GT + ++FARPNWR V+ +IA+ HP +GVFYCG P KEL+ L+
Sbjct: 210 SLNHAKEGLDILNGTPIASYFARPNWRSVYNRIANNHPQKRIGVFYCGAPAPIKELRGLA 389
Query: 920 LELSHKTTTRFDFHKEYF 937
LE S + +FDFHKE F
Sbjct: 390 LEFSQERAPKFDFHKENF 443
>BG587472 similar to GP|20160800|db putative respiratory burst oxidase
protein {Oryza sativa (japonica cultivar-group)},
partial (13%)
Length = 387
Score = 157 bits (396), Expect = 2e-38
Identities = 71/127 (55%), Positives = 97/127 (75%), Gaps = 1/127 (0%)
Frame = +3
Query: 439 TWLRSTRARKIVPFDDNINFHKIIACAIVVGVVVHAGNHLACDFPLLINSSPEEF-SLIA 497
T LR T +I+PFDDNINFHKIIA +V+G ++H G H++CDFP L++ E+F +++
Sbjct: 3 TKLRGTFLSQIIPFDDNINFHKIIAVGVVIGTLIHVGVHVSCDFPRLVSCPTEKFMAILG 182
Query: 498 SDFNNKKPTYKTLLTGVEGVTGISMVTLMAISFTLATHHFRRNAVRLPPPLNRLTGFNAF 557
S F+ K+P+Y TL+T G+TGI MV +MA SFTLATH+FR++ V LP PL+ L GFN+F
Sbjct: 183 SGFDYKQPSYLTLVTSPPGITGIFMVLIMAFSFTLATHYFRKSVVTLPSPLHHLAGFNSF 362
Query: 558 WYSHHLL 564
WY+HHLL
Sbjct: 363 WYAHHLL 383
>AW687772 similar to GP|16549089|db respiratory burst oxidase homolog
{Solanum tuberosum}, partial (13%)
Length = 622
Score = 124 bits (311), Expect = 2e-28
Identities = 67/138 (48%), Positives = 88/138 (63%), Gaps = 1/138 (0%)
Frame = +1
Query: 717 PRLFVDGPYGAPAQDFKNFDVLLLIGLGIGATPFISILRDLLNNTRASDESMDQSNTETT 776
P+L +DGPYGAPAQD+K+++V+LL+GLGIGATP ISIL+D+LNN + + ++Q E+
Sbjct: 238 PKLLIDGPYGAPAQDYKDYEVILLVGLGIGATPLISILKDVLNNMK-QQKDIEQGVVESG 414
Query: 777 RSDESWNSFTSSNLTPGGNKRSQRTTN-AYFYWVTREPGSFEWFKGVMDEVAEMDHRGQI 835
+ NKR TN AYFYWVTRE GSFEWFKGVMDE+A M +
Sbjct: 415 VKN---------------NKRKPFATNRAYFYWVTREQGSFEWFKGVMDEIAIMTRMD*L 549
Query: 836 ELHNYLTSVYEEGDARST 853
++GDARS+
Sbjct: 550 NFIIIAQVCMKKGDARSS 603
>BE203618 weakly similar to GP|3242787|gb|A respiratory burst oxidase protein
E {Arabidopsis thaliana}, partial (6%)
Length = 206
Score = 119 bits (297), Expect = 7e-27
Identities = 54/68 (79%), Positives = 64/68 (93%)
Frame = +3
Query: 467 VVGVVVHAGNHLACDFPLLINSSPEEFSLIASDFNNKKPTYKTLLTGVEGVTGISMVTLM 526
VVG++VHAGNH++CDFPLL+NSSPE+FSLIASDFNNKKPTYK+L +EGVTGI+M+TLM
Sbjct: 3 VVGILVHAGNHISCDFPLLVNSSPEKFSLIASDFNNKKPTYKSLFISIEGVTGITMLTLM 182
Query: 527 AISFTLAT 534
ISFTLAT
Sbjct: 183 VISFTLAT 206
>AW256730 weakly similar to GP|9757922|dbj respiratory burst oxidase protein
{Arabidopsis thaliana}, partial (11%)
Length = 681
Score = 94.4 bits (233), Expect = 2e-19
Identities = 54/148 (36%), Positives = 87/148 (58%), Gaps = 6/148 (4%)
Frame = +2
Query: 128 SMSMRTSASSTESVEDPVESARNARRMQ---AKLERTRSSAQRALKGLRFISKSGEAGE- 183
S ++ S + S D + S N + +K+ R S A R LKGLR + ++ E
Sbjct: 236 SKVIKASDNEAFSNVDNLRSRNNNNNINNNGSKMRRMESGAARGLKGLRVLDRTVTGKEA 415
Query: 184 ELWRKVEERFGSLAKDGLLAREDFGGCIGMD-DSKEFAVGVFDALARRKGRKASK-ITKE 241
+ W+ +E+RF A DG+L+++ FG C+GM DSK+FA +++ALARR+ A IT
Sbjct: 416 DAWKSIEKRFTQHAVDGMLSKDKFGTCMGMGADSKDFAGELYEALARRRNICAENGITLN 595
Query: 242 ELHHFWQQISDQSFDARLQIFFDMADSN 269
E FW+ ++++ +++LQ+FFDM D N
Sbjct: 596 EARVFWEDMTNKDLESKLQVFFDMCDKN 679
>BM813683 weakly similar to GP|15341529|gb ferric-chelate reductase {Pisum
sativum}, partial (21%)
Length = 523
Score = 74.3 bits (181), Expect = 2e-13
Identities = 42/138 (30%), Positives = 67/138 (48%), Gaps = 2/138 (1%)
Frame = +1
Query: 623 VLPGNVFSLIMSKPNGFKYKSGQYIFLQCPKISPFEWHPFSITSAPGDEY--LSVHIRTV 680
+LP + L SK Y +F+ PK+S +WHPF+++S+ E LSV I+ V
Sbjct: 1 LLPCDALELNFSKDPSLYYNPTSLVFINVPKVSKLQWHPFTVSSSCNLETNCLSVTIKNV 180
Query: 681 GDWTQELKLLFTEDDIPPVNCNATFGELVQMDQREHPRLFVDGPYGAPAQDFKNFDVLLL 740
G W+ +L + + +H + V+GPYG + F + + +
Sbjct: 181 GSWSNKLYQELSSSSL------------------DHLNVSVEGPYGPHSAQFLRHEQIAM 306
Query: 741 IGLGIGATPFISILRDLL 758
+ G G TPFISI+RDL+
Sbjct: 307 VSGGSGITPFISIIRDLI 360
>BE941228 similar to GP|20160800|dbj putative respiratory burst oxidase
protein {Oryza sativa (japonica cultivar-group)},
partial (5%)
Length = 392
Score = 60.1 bits (144), Expect = 4e-09
Identities = 28/50 (56%), Positives = 33/50 (66%)
Frame = +3
Query: 888 VFIKIASKHPYSTVGVFYCGMPVLAKELKKLSLELSHKTTTRFDFHKEYF 937
VF ++A+ H S +GVFYCG P L K LK L E S T+TRF FHKE F
Sbjct: 3 VFSQLATTHESSRIGVFYCGSPTLTKSLKSLCQEFSLNTSTRFHFHKENF 152
>AW585735 similar to GP|13958313|gb| NADPH oxidase 1 {Podospora anserina},
partial (4%)
Length = 496
Score = 57.8 bits (138), Expect = 2e-08
Identities = 34/103 (33%), Positives = 56/103 (54%), Gaps = 4/103 (3%)
Frame = +1
Query: 379 WVLFLWLVTTASLFAWKFYQYRNR---TSFQVMGYCLPVAKGAAETLKLNMALILLPVCR 435
W+ W + LF + F + + + V+G + ++GA L + ALILLPVCR
Sbjct: 118 WIF--WFGSHIGLFIYGFLKQKTDHELDNLNVIGLSVWSSRGAGLCLAYDGALILLPVCR 291
Query: 436 NTLTWLRSTR-ARKIVPFDDNINFHKIIACAIVVGVVVHAGNH 477
N + RS R I+PFD+N+ FH+ A A+++ ++H +H
Sbjct: 292 NLIKQFRSFRFLNNIIPFDENLWFHRQTAYAMLIFTLIHTFSH 420
>TC84288 similar to GP|17384016|emb|CAC87256. NADPH oxidase {Nicotiana
tabacum}, partial (11%)
Length = 807
Score = 52.4 bits (124), Expect = 8e-07
Identities = 45/148 (30%), Positives = 70/148 (46%), Gaps = 26/148 (17%)
Frame = +1
Query: 57 ELVEITLEL-EDDAVVLCNVAPA------SSGAGTS---------SSSSHATGDEGTSAS 100
E VE+TL+L +DD +VL +V PA SS AG+ S S + G
Sbjct: 346 EFVEVTLDLRDDDTIVLRSVEPAVINIDDSSVAGSGYDTPVSVPRSPSMRRSSSRGFRHF 525
Query: 101 GAGAVARSLSKNLSITSRIRRKFPWLRSMSMRTSASSTESVEDPVES----------ARN 150
A +++K + +RR F W + RT +SS+ + AR
Sbjct: 526 SQELKAEAVAKAKQFSQELRR-FSWSHGHASRTLSSSSARAGTSTTAGRWVLKRRWAARA 702
Query: 151 ARRMQAKLERTRSSAQRALKGLRFISKS 178
R+ +A+L+RTR+ A +AL+ L+FIS +
Sbjct: 703 LRKARAQLDRTRTGAHKALRSLKFISSN 786
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 30,873,991
Number of Sequences: 36976
Number of extensions: 464917
Number of successful extensions: 2820
Number of sequences better than 10.0: 60
Number of HSP's better than 10.0 without gapping: 2726
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2793
length of query: 937
length of database: 9,014,727
effective HSP length: 105
effective length of query: 832
effective length of database: 5,132,247
effective search space: 4270029504
effective search space used: 4270029504
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0013.6


