

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
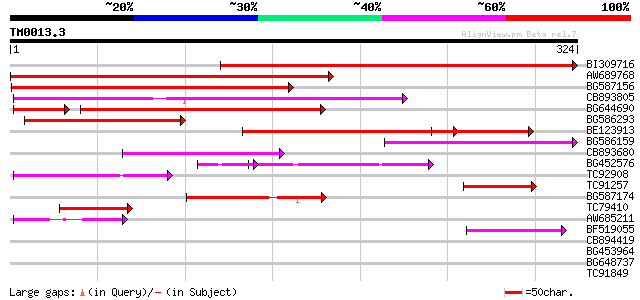
Query= TM0013.3
(324 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 234 3e-62
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 187 6e-48
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 160 5e-40
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 152 1e-37
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 123 2e-32
BG586293 weakly similar to PIR|E84473|E84 probable retroelement ... 101 3e-22
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 95 3e-20
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 86 1e-17
CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotia... 67 1e-11
BG452576 weakly similar to GP|12005223|gb|A reverse transcriptas... 57 2e-11
TC92908 similar to GP|10140704|gb|AAG13538.1 putative gag-pol po... 58 4e-09
TC91257 similar to GP|21434|emb|CAA36616.1|| ORF4 {Solanum tuber... 55 5e-08
BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse ... 50 1e-06
TC79410 similar to PIR|H84518|H84518 pathogenesis-related PR-1-l... 46 2e-05
AW685211 weakly similar to GP|4539662|gb| polyprotein {Sorghum b... 45 5e-05
BF519055 similar to GP|21592754|gb| unknown {Arabidopsis thalian... 43 2e-04
CB894419 weakly similar to GP|10177935|db copia-type polyprotein... 40 0.001
BG453964 37 0.013
BG648737 weakly similar to GP|21433|emb|CA ORF3 {Solanum tuberos... 36 0.023
TC91849 31 0.57
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 234 bits (597), Expect = 3e-62
Identities = 122/204 (59%), Positives = 150/204 (72%)
Frame = +2
Query: 121 QVCKLQKSLYGLKQASRKWYEKLSSHLRNLGYSQATSDHSLFVKFQDTLFAGLLVYVDDV 180
+VC+LQKS+YGLKQASR+WY KLS L + GY Q++SD SLF KF+D+ F LLVYVDD+
Sbjct: 17 KVCELQKSIYGLKQASRQWYSKLSESLISFGYLQSSSDFSLFTKFKDSSFTTLLVYVDDI 196
Query: 181 ILFGNCLAEFQVVKDSLHTAFGIKDLVILKYFLGLEVAHSSLGISLCQRKYCLDLLTETG 240
+L GN ++E Q VK L F IKDL L+YFLGLEVA S GI L QRKY L+LL ++G
Sbjct: 197 VLAGNDISEIQHVKCFLIDRFKIKDLGSLRYFLGLEVARSKQGILLNQRKYTLELLEDSG 376
Query: 241 TLGSKPVSTPLDPSCKLVVNQGEPYSDATAYRRLVGRLLYLTTTRPDISFATQQLSQFMS 300
L K TP D S KL + Y+D T YRRL+G+L+YLTTTRPDISFA QQLSQF+S
Sbjct: 377 NLAVKSTLTPYDISLKLHNSDSPLYNDETQYRRLIGKLIYLTTTRPDISFAVQQLSQFVS 556
Query: 301 NPMTDHYKAALRVLRFLKTSPGLG 324
P HY+AA+RVL++LKT+P G
Sbjct: 557 KPQQVHYQAAIRVLQYLKTAPAKG 628
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 187 bits (474), Expect = 6e-48
Identities = 90/185 (48%), Positives = 124/185 (66%)
Frame = +1
Query: 1 KAEIAALEKNETWKIVDLPQGVKAIGNKWVYRIKRNADGTLARYKARLVAKGYNQIEGLD 60
K E AL N+TW +V LP KAIG KWVYR+K N DG++ ++KARLVAKG++Q G D
Sbjct: 112 KTEYKALIDNKTWDLVPLPPHKKAIGCKWVYRVKENPDGSVNKFKARLVAKGFSQTLGCD 291
Query: 61 YFDTFSPVAKLTTVRMVLALAAAQNWHLHQLDVDNAFLHGTLDEDVYMNIPAGVPTMKPN 120
Y +TFSPV K T+R++L +A W + Q+D++NAFL+G L E+VYM+ P G +
Sbjct: 292 YTETFSPVIKPVTIRLILTIAITYKWEIQQIDINNAFLNGFLQEEVYMSQPQGFEAANKS 471
Query: 121 QVCKLQKSLYGLKQASRKWYEKLSSHLRNLGYSQATSDHSLFVKFQDTLFAGLLVYVDDV 180
VCKL KSLYGLKQA R WYE L+S G++++ D SL + Q+ L +YVDD+
Sbjct: 472 LVCKLNKSLYGLKQAPRAWYEXLTSAQIQFGFTKSRCDPSLLIYNQNGACIYLXIYVDDI 651
Query: 181 ILFGN 185
++ G+
Sbjct: 652 LITGS 666
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 160 bits (406), Expect = 5e-40
Identities = 78/162 (48%), Positives = 112/162 (68%), Gaps = 1/162 (0%)
Frame = -1
Query: 2 AEIAALEKNETWKIVDLPQGVKAIGNKWVYRIKRNADGTLARYKARLVAKGYNQIEGLDY 61
AE A+ KN+TW +LP+G KA+ ++W++ IK ADG++ R K RLVA+G+ G DY
Sbjct: 504 AEAGAMIKNDTWYESELPKGKKAVSSRWIFTIKYKADGSIERKKTRLVARGFTLTYGEDY 325
Query: 62 FDTFSPVAKLTTVRMVLALAAAQNWHLHQLDVDNAFLHGTLDEDVYMNIPAGVP-TMKPN 120
+TF+PVAKL T+R+VL+LA W L Q+DV NAFL G L+++VYM P G+ +K
Sbjct: 324 IETFAPVAKLHTIRIVLSLAVNLGWGLWQMDVKNAFLQGELEDEVYMYPPPGLEHLVKRG 145
Query: 121 QVCKLQKSLYGLKQASRKWYEKLSSHLRNLGYSQATSDHSLF 162
V +L+K++YGLKQ+ R WY KLS+ L G+ ++ DH+LF
Sbjct: 144 NVLRLKKAIYGLKQSPRAWYNKLSTTLNGRGFRKSELDHTLF 19
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 152 bits (385), Expect = 1e-37
Identities = 85/233 (36%), Positives = 133/233 (56%), Gaps = 8/233 (3%)
Frame = +3
Query: 3 EIAALEKNETWKIVDLPQGVKAIGNKWVYRIKRNADGTLARYKARLVAKGYNQIEGLDYF 62
E+ A E+N TW++ DL G K IG KW+++ K N +G + +YKARLVAKGY+Q G+DY
Sbjct: 99 EMEATERNNTWELTDLRSGAKTIGLKWIFKTKLNENGEIEKYKARLVAKGYSQQYGVDYT 278
Query: 63 DTFSPVAKLTTVRMVLALAAAQNWHLHQLDVDNAFL------HGTLDEDVYMNIPAGVPT 116
+ F+PVA+ T+RMV+ALAA Q+ D + H ++ + +
Sbjct: 279 EVFAPVARWDTIRMVIALAA-------QIKRDGVCIS*M*KAHSCMEN*MRKFLLINHRV 437
Query: 117 MKPNQVC-KLQKSLYGLKQASRKWYEKLSSHLRNLGYSQATSDHSLFVKFQD-TLFAGLL 174
M + +++++LYGLKQA R WY ++ ++ G+ + +H+LFVK + +
Sbjct: 438 M*RRVIS*RVKRALYGLKQAPRAWYSRIEAYFTKEGFEKCPYEHTLFVKLSEGGKILIIS 617
Query: 175 VYVDDVILFGNCLAEFQVVKDSLHTAFGIKDLVILKYFLGLEVAHSSLGISLC 227
+YVDD+I GN F+ K S+ F + DL + YFLG+EV + GI +C
Sbjct: 618 LYVDDLIFIGNDENMFEEFKKSMKKEFNMSDLGKMHYFLGVEVTQNEKGIYIC 776
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 123 bits (309), Expect(2) = 2e-32
Identities = 61/141 (43%), Positives = 95/141 (67%), Gaps = 1/141 (0%)
Frame = -2
Query: 41 LARYKARLVAKGYNQIEGLDYFDTFSPVAKLTTVRMVLALAAAQNWHLHQLDVDNAFLHG 100
+ R K++LV +GYNQ EG+DY + FSPVA++ +R+++A AA + L+Q+DV +AF++G
Sbjct: 424 ITRNKSKLVVQGYNQKEGIDYDEAFSPVARMEAIRILIAFAAFMGFKLYQMDVKSAFING 245
Query: 101 TLDEDVYMNIPAGVPTMK-PNQVCKLQKSLYGLKQASRKWYEKLSSHLRNLGYSQATSDH 159
L E+V++ P G + PN V +L K+LYGLKQA R WYE+LS L G+ + D+
Sbjct: 244 DLKEEVFVKQPPGFEDAEVPNHVFRLNKTLYGLKQAPRAWYERLSKFLLKNGFKRGKIDN 65
Query: 160 SLFVKFQDTLFAGLLVYVDDV 180
+LF+ ++ + VYVDD+
Sbjct: 64 TLFLLKRE*ELLIIQVYVDDI 2
Score = 33.1 bits (74), Expect(2) = 2e-32
Identities = 12/32 (37%), Positives = 20/32 (62%)
Frame = -3
Query: 3 EIAALEKNETWKIVDLPQGVKAIGNKWVYRIK 34
E+ E+++ W +V P+G IG +WV+R K
Sbjct: 528 ELHQFERSKVWYLVPRPEGKTVIGTRWVFRNK 433
>BG586293 weakly similar to PIR|E84473|E84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 763
Score = 101 bits (252), Expect = 3e-22
Identities = 47/92 (51%), Positives = 68/92 (73%)
Frame = +2
Query: 9 KNETWKIVDLPQGVKAIGNKWVYRIKRNADGTLARYKARLVAKGYNQIEGLDYFDTFSPV 68
+ +T K+V P GVK IG +W+Y+IKRN DGTL +YKARLVAKGY + +G+D+ + F+PV
Sbjct: 47 QKQTLKLVKKPTGVKPIGLRWIYKIKRNEDGTLIKYKARLVAKGYVKQQGIDFDEVFAPV 226
Query: 69 AKLTTVRMVLALAAAQNWHLHQLDVDNAFLHG 100
++ T+ ++LALAA +H +DV AFL+G
Sbjct: 227 VRIETI*LLLALAATNGC*IHHIDVKIAFLNG 322
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 95.1 bits (235), Expect = 3e-20
Identities = 52/124 (41%), Positives = 76/124 (60%), Gaps = 1/124 (0%)
Frame = +1
Query: 134 QASRKWYEKLSSHLRNLGYSQATSDHSLFVKFQDTLF-AGLLVYVDDVILFGNCLAEFQV 192
Q+ R W+++ + ++ GY Q +DH++F+K T+ A L+VYVDD+ L G+ +
Sbjct: 1 QSPRDWFDRFT*VVKKFGYIQCQTDHAMFIKHSSTVKKAILIVYVDDIFLTGDHGK*IKR 180
Query: 193 VKDSLHTAFGIKDLVILKYFLGLEVAHSSLGISLCQRKYCLDLLTETGTLGSKPVSTPLD 252
+K+ L F IKDL LKYFLG+EVA G S+ QRKY LDLL ET +G K + P
Sbjct: 181 LKNLLAEEFEIKDLGNLKYFLGMEVARWKKGSSISQRKYVLDLLKETRMIGCKTIRDPYG 360
Query: 253 PSCK 256
+C+
Sbjct: 361 CNCE 372
Score = 54.3 bits (129), Expect = 6e-08
Identities = 28/58 (48%), Positives = 35/58 (60%)
Frame = +2
Query: 242 LGSKPVSTPLDPSCKLVVNQGEPYSDATAYRRLVGRLLYLTTTRPDISFATQQLSQFM 299
L KP TP+D + KL D Y+RLVG+L+YL+ TRPDISF +SQFM
Sbjct: 329 LDVKPSETPMDATVKLGTLDNGTLVDKGRYQRLVGKLIYLSHTRPDISFVVCTMSQFM 502
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 86.3 bits (212), Expect = 1e-17
Identities = 45/110 (40%), Positives = 63/110 (56%)
Frame = +1
Query: 215 LEVAHSSLGISLCQRKYCLDLLTETGTLGSKPVSTPLDPSCKLVVNQGEPYSDATAYRRL 274
+EV + GI +CQRKY DLL G S P+ P CKL+ ++ DAT Y+++
Sbjct: 1 VEVIQNEEGIYICQRKYVTDLLERFGMEKSNLSRNPIAPRCKLIKDENGVKVDATKYKQI 180
Query: 275 VGRLLYLTTTRPDISFATQQLSQFMSNPMTDHYKAALRVLRFLKTSPGLG 324
VG L+YL TRPD+ + +S+FM+ P H A RVLR+L + LG
Sbjct: 181 VGCLMYLAATRPDLMYVLSLISRFMNCPTELHMHAVKRVLRYLNGTINLG 330
>CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotiana tabacum},
partial (7%)
Length = 780
Score = 66.6 bits (161), Expect = 1e-11
Identities = 37/93 (39%), Positives = 51/93 (54%)
Frame = -2
Query: 65 FSPVAKLTTVRMVLALAAAQNWHLHQLDVDNAFLHGTLDEDVYMNIPAGVPTMKPNQVCK 124
F P+ KL T+ +L++ A +N +L LDV AFL G L ED+YM+ P G V K
Sbjct: 554 FVPIVKLNTIMFLLSIVAIENLYLE*LDVKTAFLRGDLVEDIYMHQPEGFS*EVGKMVGK 375
Query: 125 LQKSLYGLKQASRKWYEKLSSHLRNLGYSQATS 157
L+KS+YGLKQ R+ L + G+ S
Sbjct: 374 LKKSMYGLKQGPRQCI*SLKALCTRKGFRSEIS 276
>BG452576 weakly similar to GP|12005223|gb|A reverse transcriptase-like
protein {Spiranthes hongkongensis}, partial (25%)
Length = 676
Score = 57.0 bits (136), Expect(2) = 2e-11
Identities = 43/106 (40%), Positives = 60/106 (56%)
Frame = +2
Query: 137 RKWYEKLSSHLRNLGYSQATSDHSLFVKFQDTLFAGLLVYVDDVILFGNCLAEFQVVKDS 196
RK Y KL S L +LGY Q+ +DHS+F F LVY+DD++ + +E Q+VK
Sbjct: 236 RK*Y*KLLSTLISLGYKQSPNDHSIF--SFGRRFTIFLVYLDDIVFARDDHSETQLVKSH 409
Query: 197 LHTAFGIKDLVILKYFLGLEVAHSSLGISLCQRKYCLDLLTETGTL 242
L F I L L Y +G+++A S I Q KY ++LL E+G L
Sbjct: 410 LDKNFIIIGLGTLHY-VGVKIA*SES*IIDDQCKYTIELLEESGQL 544
Score = 29.3 bits (64), Expect(2) = 2e-11
Identities = 14/35 (40%), Positives = 20/35 (57%)
Frame = +3
Query: 108 MNIPAGVPTMKPNQVCKLQKSLYGLKQASRKWYEK 142
+N+ +P + VCK S+YGLK A R+ Y K
Sbjct: 129 VNVLGSIPCLIA--VCKFHNSIYGLKHAHRQ*YSK 227
>TC92908 similar to GP|10140704|gb|AAG13538.1 putative gag-pol polyprotein
{Oryza sativa}, partial (1%)
Length = 638
Score = 58.2 bits (139), Expect = 4e-09
Identities = 36/91 (39%), Positives = 53/91 (57%)
Frame = +2
Query: 3 EIAALEKNETWKIVDLPQGVKAIGNKWVYRIKRNADGTLARYKARLVAKGYNQIEGLDYF 62
EI LE+N T + L +G K + + VY+I A+G + +YKA+LVAK + Q+EG D+
Sbjct: 338 EIQVLEENNTSTLEPLREGKKWVDCRPVYKIIHKANGEIEKYKAQLVAKDFVQVEGEDF* 517
Query: 63 DTFSPVAKLTTVRMVLALAAAQNWHLHQLDV 93
D K R +L +AAA+ LH +DV
Sbjct: 518 D-LCLSNKDDNCRCLLTIAAAKG*QLHLMDV 607
>TC91257 similar to GP|21434|emb|CAA36616.1|| ORF4 {Solanum tuberosum},
partial (8%)
Length = 854
Score = 54.7 bits (130), Expect = 5e-08
Identities = 25/42 (59%), Positives = 33/42 (78%)
Frame = +3
Query: 260 NQGEPYSDATAYRRLVGRLLYLTTTRPDISFATQQLSQFMSN 301
+QGE +SD YRRLVG+L YLT TRPDIS+A +SQF+++
Sbjct: 729 DQGETFSDPGRYRRLVGKLNYLTMTRPDISYAVSVVSQFLNS 854
>BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse
transcriptase homolog - rape retrotransposon copia-like
(fragment), partial (84%)
Length = 249
Score = 50.1 bits (118), Expect = 1e-06
Identities = 32/86 (37%), Positives = 52/86 (60%), Gaps = 6/86 (6%)
Frame = -1
Query: 102 LDEDVYMNIPAG-VPTMKPNQVCKLQKSLYGLKQASRKWYEKLSSHLRNLGYSQATSDHS 160
L+E +YM P G + K + VCKL+KSLYGLKQ+ R+WY++ S+ S AT+
Sbjct: 246 LEEKIYMTQPEGFLFPGKEDHVCKLRKSLYGLKQSPRQWYKRFDSYRS----SWATTGVL 79
Query: 161 LFV-----KFQDTLFAGLLVYVDDVI 181
+ V + + + + L++YVDD++
Sbjct: 78 MTVVST*TR*RMSRYIYLVLYVDDML 1
>TC79410 similar to PIR|H84518|H84518 pathogenesis-related PR-1-like protein
[imported] - Arabidopsis thaliana, partial (84%)
Length = 1002
Score = 46.2 bits (108), Expect = 2e-05
Identities = 21/42 (50%), Positives = 29/42 (69%)
Frame = +3
Query: 29 WVYRIKRNADGTLARYKARLVAKGYNQIEGLDYFDTFSPVAK 70
+ + +KRN+DG+ Y RLVAKG++Q G+DY D FS V K
Sbjct: 756 YFFCLKRNSDGSTLYYTTRLVAKGFHQRSGIDYKDQFSLVVK 881
>AW685211 weakly similar to GP|4539662|gb| polyprotein {Sorghum bicolor},
partial (2%)
Length = 388
Score = 44.7 bits (104), Expect = 5e-05
Identities = 26/65 (40%), Positives = 34/65 (52%)
Frame = -2
Query: 3 EIAALEKNETWKIVDLPQGVKAIGNKWVYRIKRNADGTLARYKARLVAKGYNQIEGLDYF 62
E+ ALE N TW I+ LP + Y+ R K R+ AKGY+++ GL Y
Sbjct: 207 EMTALESNRTWTIIPLPLEMP-------YK---------HRLKVRMAAKGYSKVYGLHYN 76
Query: 63 DTFSP 67
DTFSP
Sbjct: 75 DTFSP 61
>BF519055 similar to GP|21592754|gb| unknown {Arabidopsis thaliana}, partial
(30%)
Length = 675
Score = 42.7 bits (99), Expect = 2e-04
Identities = 21/57 (36%), Positives = 33/57 (57%)
Frame = -3
Query: 262 GEPYSDATAYRRLVGRLLYLTTTRPDISFATQQLSQFMSNPMTDHYKAALRVLRFLK 318
G +D+ +VG LLY+T T PD+SF+ + SQFM P +++ +V+R K
Sbjct: 661 GSITADSKLVHSIVGALLYITVTCPDLSFSINKPSQFMHKPTQINFQQLKKVMRHPK 491
>CB894419 weakly similar to GP|10177935|db copia-type polyprotein
{Arabidopsis thaliana}, partial (2%)
Length = 170
Score = 40.0 bits (92), Expect = 0.001
Identities = 21/56 (37%), Positives = 30/56 (53%)
Frame = +3
Query: 229 RKYCLDLLTETGTLGSKPVSTPLDPSCKLVVNQGEPYSDATAYRRLVGRLLYLTTT 284
+K D+L + SKP+STP++ KL D+T Y+ L+G L YLT T
Sbjct: 3 KKCASDILKKFKMEHSKPISTPVEEKLKLTRESDGKRVDSTHYKSLIGSLRYLTAT 170
>BG453964
Length = 647
Score = 36.6 bits (83), Expect = 0.013
Identities = 21/62 (33%), Positives = 33/62 (52%)
Frame = -2
Query: 170 FAGLLVYVDDVILFGNCLAEFQVVKDSLHTAFGIKDLVILKYFLGLEVAHSSLGISLCQR 229
F LLVY+D+++L + ++ L F KDL + KYF G+ VA S+ + +
Sbjct: 205 FIYLLVYIDEILL-SIVIIPLVCLRQHLSNHFQTKDLDLFKYFSGIVVAQSTTHATTTKL 29
Query: 230 KY 231
KY
Sbjct: 28 KY 23
>BG648737 weakly similar to GP|21433|emb|CA ORF3 {Solanum tuberosum}, partial
(10%)
Length = 804
Score = 35.8 bits (81), Expect = 0.023
Identities = 16/26 (61%), Positives = 19/26 (72%)
Frame = +3
Query: 3 EIAALEKNETWKIVDLPQGVKAIGNK 28
EI ALEKNETW++ DLP G +G K
Sbjct: 726 EIHALEKNETWELSDLPSGKHPMGCK 803
>TC91849
Length = 702
Score = 31.2 bits (69), Expect = 0.57
Identities = 22/66 (33%), Positives = 31/66 (46%)
Frame = +3
Query: 197 LHTAFGIKDLVILKYFLGLEVAHSSLGISLCQRKYCLDLLTETGTLGSKPVSTPLDPSCK 256
L++ F I+DL LKY++GLE++ S T+T L SK ST + K
Sbjct: 546 LNSWFKIEDLG*LKYYVGLEISRSQ---------------TDTILLASKSASTRIQRGTK 680
Query: 257 LVVNQG 262
L G
Sbjct: 681 LPYESG 698
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.137 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,375,072
Number of Sequences: 36976
Number of extensions: 122143
Number of successful extensions: 521
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 511
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 514
length of query: 324
length of database: 9,014,727
effective HSP length: 96
effective length of query: 228
effective length of database: 5,465,031
effective search space: 1246027068
effective search space used: 1246027068
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0013.3


