

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
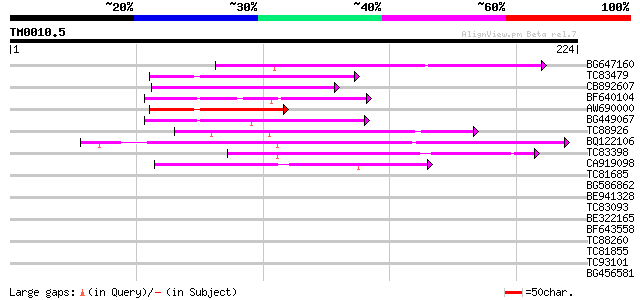
Query= TM0010.5
(224 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG647160 homologue to GP|15004435|gb| maturase {Ionopsis satyrio... 60 9e-10
TC83479 similar to GP|23476992|emb|CAD48949. hypothetical protei... 59 1e-09
CB892607 similar to GP|8927657|gb| EST gb|N38213 comes from this... 59 2e-09
BF640104 51 4e-07
AW690000 51 4e-07
BG449067 49 2e-06
TC88926 similar to GP|7290507|gb|AAF45960.1| peb gene product {D... 46 1e-05
BQ122106 similar to GP|9366656|emb|C probable similar to ring-h2... 45 3e-05
TC83398 similar to PIR|T05150|T05150 hypothetical protein F18E5.... 44 5e-05
CA919098 42 1e-04
TC81685 40 0.001
BG586862 38 0.003
BE941328 37 0.008
TC83093 35 0.023
BE322165 35 0.030
BF643558 34 0.039
TC88260 homologue to GP|15215696|gb|AAK91394.1 AT3g16950/K14A17_... 33 0.087
TC81855 homologue to GP|15290186|dbj|BAB63876. putative lipoamid... 33 0.11
TC93101 31 0.33
BG456581 31 0.33
>BG647160 homologue to GP|15004435|gb| maturase {Ionopsis satyrioides},
partial (2%)
Length = 780
Score = 59.7 bits (143), Expect = 9e-10
Identities = 40/132 (30%), Positives = 64/132 (48%), Gaps = 1/132 (0%)
Frame = +1
Query: 82 GGVLRDHNGNYLVGFSALLGRC-SINFAELWGIYHGAKLALDRGFKKVMIESDSAFAVNL 140
GGV+R++ Y+ GFS + +I FAEL ++ KLA+ ++++ SDS VNL
Sbjct: 379 GGVIRNYLSTYITGFSGFISIYQNIMFAELTTLHQSLKLAISLNIEEMVCYSDSLLTVNL 558
Query: 141 VNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHVFREINSVADAFAKNGLGLLMDFRIYDA 200
+ + A L+ NIK +S+ L H RE N AD K +D I+ +
Sbjct: 559 IKEDISQHHVYAVLIHNIKYIMSS-RNFTLHHTLREGNQCADFMVKLRTSTDVDLTIHSS 735
Query: 201 IPSFISVALMHD 212
P + ++ D
Sbjct: 736 PPKVLLPSVRTD 771
>TC83479 similar to GP|23476992|emb|CAD48949. hypothetical protein {Plasmodium
falciparum 3D7}, partial (0%)
Length = 1222
Score = 59.3 bits (142), Expect = 1e-09
Identities = 31/83 (37%), Positives = 42/83 (50%)
Frame = +2
Query: 56 WVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGIYH 115
W P W K+NTDGS + A GG+LRD+ G + F + + ELW I+
Sbjct: 977 WKKPEIGWTKLNTDGSVN--KETAGFGGLLRDYRGEPICAFVSKAPQGDTFLVELWAIWR 1150
Query: 116 GAKLALDRGFKKVMIESDSAFAV 138
G L+L G K + +ESDS V
Sbjct: 1151 GLVLSLGLGIKSIWVESDSMSVV 1219
>CB892607 similar to GP|8927657|gb| EST gb|N38213 comes from this gene.
{Arabidopsis thaliana}, partial (1%)
Length = 782
Score = 58.5 bits (140), Expect = 2e-09
Identities = 26/74 (35%), Positives = 43/74 (57%)
Frame = +1
Query: 57 VPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGIYHG 116
+PPN + + D AACGG+++D G+++ ++ LG CS+ AELWGI HG
Sbjct: 556 IPPNSCEVALKCDYVVLNCDLNAACGGLIQDDQGHFVFHYANKLGSCSVLQAELWGI*HG 735
Query: 117 AKLALDRGFKKVMI 130
+ +RG+ K+ +
Sbjct: 736 LSIDWNRGYSKIRV 777
>BF640104
Length = 344
Score = 50.8 bits (120), Expect = 4e-07
Identities = 30/94 (31%), Positives = 53/94 (55%), Gaps = 4/94 (4%)
Frame = -1
Query: 54 LRWVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGR----CSINFAE 109
++W+ P+ IKIN D + + ++ G + R+ NG +V S R C I AE
Sbjct: 344 VKWIKPHQGVIKINCDANLTS-EDVWGIGVITRNDNG--IVMASGTWNRPGFMCPIT-AE 177
Query: 110 LWGIYHGAKLALDRGFKKVMIESDSAFAVNLVNK 143
WG+Y A ALD+GF+ V+ E+D+ ++++++
Sbjct: 176 AWGVYQAALFALDQGFQNVLFENDNEKLISMLSR 75
>AW690000
Length = 652
Score = 50.8 bits (120), Expect = 4e-07
Identities = 25/55 (45%), Positives = 34/55 (61%)
Frame = +3
Query: 56 WVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAEL 110
W PP WIK NTDGS S+ +ACGG+ R+H+ + L+ F+ G C+ AEL
Sbjct: 411 WRPPIPHWIKCNTDGSSR--SHSSACGGIFRNHDTDLLLCFAENTGECNAFHAEL 569
>BG449067
Length = 578
Score = 48.5 bits (114), Expect = 2e-06
Identities = 31/90 (34%), Positives = 45/90 (49%), Gaps = 1/90 (1%)
Frame = -2
Query: 54 LRWVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLV-GFSALLGRCSINFAELWG 112
++W P D IK+N+D + S ++L G V R+ G + G G S AE WG
Sbjct: 313 VKWKKPEKDIIKLNSDANLSS-TDLWGIGVVARNDEGFAMASGTWFRFGFPSATTAEAWG 137
Query: 113 IYHGAKLALDRGFKKVMIESDSAFAVNLVN 142
IY A + GF KV ESD+ + ++N
Sbjct: 136 IYQAMIFAGEYGFSKVQFESDNERVIQMLN 47
>TC88926 similar to GP|7290507|gb|AAF45960.1| peb gene product {Drosophila
melanogaster}, partial (1%)
Length = 1073
Score = 46.2 bits (108), Expect = 1e-05
Identities = 35/123 (28%), Positives = 58/123 (46%), Gaps = 3/123 (2%)
Frame = +3
Query: 66 INTDGSYSPVSNL--AACGGVLRDHNGNYLVGFSALLG-RCSINFAELWGIYHGAKLALD 122
+N DGS + A CGGVL D +G +L GF+ L ++ E I G +
Sbjct: 348 LNVDGSLLREREVPSAGCGGVLSDSSGKWLCGFAQKLNPNLKVDETEKEAILRGLLWVKE 527
Query: 123 RGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHVFREINSVAD 182
+G +K++++SD+ V VN G + P ++++ S + L + N+VAD
Sbjct: 528 KGKRKILVKSDNEGVVYSVNCGGRSNDPLVCGIRDLLN--SPHWEATLTCIHGRSNAVAD 701
Query: 183 AFA 185
A
Sbjct: 702 RLA 710
>BQ122106 similar to GP|9366656|emb|C probable similar to ring-h2 finger
protein rha1a. {Trypanosoma brucei}, partial (18%)
Length = 693
Score = 44.7 bits (104), Expect = 3e-05
Identities = 50/195 (25%), Positives = 80/195 (40%), Gaps = 2/195 (1%)
Frame = -2
Query: 29 SQDFLY-TLTDATDDALIQRAHGTGDLRWVPPNDDWIKINTDGSYSPVSNLAACGGVLRD 87
+Q FLY +L+ ATD + +W N+ +N DGS G++R+
Sbjct: 623 AQPFLYNSLSQATDTVV----------KWNCDNNSCYILNVDGSCLGNP*PTGFNGLIRN 474
Query: 88 HNGNYLVGFSALLGRCS-INFAELWGIYHGAKLALDRGFKKVMIESDSAFAVNLVNKGCL 146
G + GF + S I AEL I+ G ++ D G + DS V+L+N +
Sbjct: 473 IAGLFNSGFPGNITNTSDILLAELHAIFQGLRMISDMGISDFVCYFDSLHYVSLINGPSM 294
Query: 147 PSKPAAALVKNIKRTLSNFDQVHLKHVFREINSVADAFAKNGLGLLMDFRIYDAIPSFIS 206
A L+++IK L + + H E N AD G I+ + P +
Sbjct: 293 KFHVYATLIQDIK-DLVITSKASVFHTLCEGNYCADFLEMLGAASDSVLTIHVSPPDGMI 117
Query: 207 VALMHDSCNTLYHRG 221
+ + TL+ RG
Sbjct: 116 QLIKVGAWETLFSRG 72
>TC83398 similar to PIR|T05150|T05150 hypothetical protein F18E5.40 -
Arabidopsis thaliana, partial (6%)
Length = 766
Score = 43.9 bits (102), Expect = 5e-05
Identities = 40/124 (32%), Positives = 59/124 (47%), Gaps = 1/124 (0%)
Frame = +2
Query: 87 DHNGNYLVGFSALLGRCS-INFAELWGIYHGAKLALDRGFKKVMIESDSAFAVNLVNKGC 145
+H G + GFS + + I FAEL I G +LA V+ SDS VNL+N
Sbjct: 80 EHFGFFNSGFSGHIDHSNDILFAELHAILMGLQLAQTLNIVDVVCYSDSLHYVNLINGPS 259
Query: 146 LPSKPAAALVKNIKRTLSNFDQVHLKHVFREINSVADAFAKNGLGLLMDFRIYDAIPSFI 205
+ A L+++IK + ++ H RE N AD AK G + I+ A P +I
Sbjct: 260 VVYHAYATLIQDIKDLI----RLSKLHTLREGNRCADFLAKLGASVDSTLPIH-ATPLWI 424
Query: 206 SVAL 209
+ +L
Sbjct: 425 ASSL 436
>CA919098
Length = 776
Score = 42.4 bits (98), Expect = 1e-04
Identities = 33/114 (28%), Positives = 49/114 (42%), Gaps = 4/114 (3%)
Frame = +2
Query: 58 PPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGIYHGA 117
P +DW+ +N D + S ++ AACG VL DH GN + +S+ + C + W Y +
Sbjct: 263 PFYEDWLMLNCDVAMSHLAGKAACGCVLHDHVGNLIFRYSSDVSLCQLP----WQNYELS 430
Query: 118 KLALDRGFKKVMIESDSAF----AVNLVNKGCLPSKPAAALVKNIKRTLSNFDQ 167
AL ++ F L+ KGC+ S LV I L Q
Sbjct: 431 SKALIF*LTYRVLTK*GLF*TLLLWCLILKGCISSHHCFLLVT*ISNLLDRLSQ 592
>TC81685
Length = 1706
Score = 39.7 bits (91), Expect = 0.001
Identities = 20/45 (44%), Positives = 24/45 (52%)
Frame = -2
Query: 56 WVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALL 100
W P DW KIN+DG S A CGG++ G +L GFS L
Sbjct: 172 WTLPQSDWDKINSDGMCQG-SLRAGCGGLI*GDGGEWLCGFSKFL 41
>BG586862
Length = 804
Score = 38.1 bits (87), Expect = 0.003
Identities = 40/135 (29%), Positives = 51/135 (37%), Gaps = 2/135 (1%)
Frame = -1
Query: 2 IWTARNELVFEKLGHTPAVVCSKIKKLSQDFLYTLTDATDDALIQRAHGT-GDLRWVPPN 60
IW ARN+ VFE + D D +IQRA + + +
Sbjct: 339 IWHARNQKVFENI-----------------------DVPGDVVIQRASSSLHSFKMAQVS 229
Query: 61 DDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALL-GRCSINFAELWGIYHGAKL 119
D + N SYS L G V R+ G + + L G AE WGIY
Sbjct: 228 DSVLPSNAIPSYS----LWGIGVVARNCEGLAMASGTWLRHGIPCATTAEAWGIYQAMVF 61
Query: 120 ALDRGFKKVMIESDS 134
A D GF K ESD+
Sbjct: 60 AGDCGFSKFEFESDN 16
>BE941328
Length = 505
Score = 36.6 bits (83), Expect = 0.008
Identities = 22/73 (30%), Positives = 36/73 (49%), Gaps = 1/73 (1%)
Frame = +3
Query: 82 GGVLRDHNGNYLVGFSALLGRCS-INFAELWGIYHGAKLALDRGFKKVMIESDSAFAVNL 140
GG+LR+ ++ FS + + I A+L I+ G + +D VM SDS AVNL
Sbjct: 9 GGILRNSASLFISSFSGFIPNSTDIL*AKLTAIHQGLNMIIDLNMNDVMCYSDSLLAVNL 188
Query: 141 VNKGCLPSKPAAA 153
+ + P ++
Sbjct: 189 IMNDTRDTTPTSS 227
>TC83093
Length = 477
Score = 35.0 bits (79), Expect = 0.023
Identities = 37/155 (23%), Positives = 66/155 (41%), Gaps = 14/155 (9%)
Frame = -3
Query: 2 IWTARNELVFE----------KLGHTPA---VVCSKIKKLSQDFLYTLTDATDDALIQRA 48
IW +RN+L+F K +T + C I K S T T T L +
Sbjct: 472 IWFSRNKLIFNNNFSSEEDTIKQSNTNIQDYITCDSINKTST--CMTQTGTTTQTLSRNV 299
Query: 49 HGTGDLRWVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLV-GFSALLGRCSINF 107
+ PP+++ IK+ + + S N + R++ G+ L L G +
Sbjct: 298 Y*E------PPSNNTIKVKINANLSK-ENTWGLSAICRNNFGDVLATAIWCLPGTHNSAA 140
Query: 108 AELWGIYHGAKLALDRGFKKVMIESDSAFAVNLVN 142
E + IY +LA +GF ++ +ESD+ + +++
Sbjct: 139 VEAFAIYLAMELADKKGFHELELESDNEQIIRILS 35
>BE322165
Length = 650
Score = 34.7 bits (78), Expect = 0.030
Identities = 16/39 (41%), Positives = 21/39 (53%)
Frame = -3
Query: 104 SINFAELWGIYHGAKLALDRGFKKVMIESDSAFAVNLVN 142
S AE WGIY A + GF KV ESD+ + ++N
Sbjct: 171 SATTAEAWGIYQAMIFAGEYGFSKVQFESDNERVIQMLN 55
>BF643558
Length = 645
Score = 34.3 bits (77), Expect = 0.039
Identities = 15/31 (48%), Positives = 21/31 (67%)
Frame = +2
Query: 53 DLRWVPPNDDWIKINTDGSYSPVSNLAACGG 83
++ W PP WIK NTDGS + +N ++CGG
Sbjct: 548 EVLWNPPPLHWIKCNTDGSSN--TNTSSCGG 634
>TC88260 homologue to GP|15215696|gb|AAK91394.1 AT3g16950/K14A17_7
{Arabidopsis thaliana}, partial (80%)
Length = 2274
Score = 33.1 bits (74), Expect = 0.087
Identities = 24/66 (36%), Positives = 36/66 (54%), Gaps = 1/66 (1%)
Frame = +3
Query: 115 HGAKL-ALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHV 173
HGA L A+++G K ++E D VN+GC+PSK A V R L N HLK +
Sbjct: 339 HGAALHAVEKGLKTAIVEGD-VVGGTCVNRGCVPSK-ALLAVSGRMRDLRN--DHHLKSL 506
Query: 174 FREINS 179
++++
Sbjct: 507 GIQVSN 524
>TC81855 homologue to GP|15290186|dbj|BAB63876. putative lipoamide
dehydrogenase {Oryza sativa (japonica cultivar-group)},
partial (13%)
Length = 591
Score = 32.7 bits (73), Expect = 0.11
Identities = 18/51 (35%), Positives = 29/51 (56%), Gaps = 1/51 (1%)
Frame = +2
Query: 115 HGAKL-ALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSN 164
HGA L A+++G K ++E D VN+GC+PSK A+ ++ S+
Sbjct: 437 HGAALHAVEKGLKTAIVEGD-VVGGTCVNRGCVPSKALLAVSGRMRELKSD 586
>TC93101
Length = 675
Score = 31.2 bits (69), Expect = 0.33
Identities = 33/138 (23%), Positives = 54/138 (38%), Gaps = 13/138 (9%)
Frame = -1
Query: 2 IWTARNELVFEKLGHTPAVVC------------SKIKKLSQDFLYTLTDATDDALIQRAH 49
IW ARN+ +FE + + + IK + + + + AT + R
Sbjct: 411 IWHARNKAIFENQFVSEDTIIQ*AQNSILAYEQATIKPQNPNIVLSSLSATSNTNTTRRR 232
Query: 50 GTGDLRWVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFS-ALLGRCSINFA 108
RW P ++ +K N D + V G ++R+ +G V + + G A
Sbjct: 231 SNVRSRWQKPLNNILKANCDANLQ-VQGRWGLGCIIRNADGEAKVTATWCINGFDCAATA 55
Query: 109 ELWGIYHGAKLALDRGFK 126
E + I LA D GFK
Sbjct: 54 ETYAILAAMYLAKDYGFK 1
>BG456581
Length = 683
Score = 31.2 bits (69), Expect = 0.33
Identities = 14/28 (50%), Positives = 18/28 (64%)
Frame = +2
Query: 56 WVPPNDDWIKINTDGSYSPVSNLAACGG 83
W PP+ W+K NTDGS ++N A GG
Sbjct: 602 WKPPSAPWVKGNTDGSX--LNNSGAXGG 679
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.139 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,496,876
Number of Sequences: 36976
Number of extensions: 98216
Number of successful extensions: 393
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 385
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 386
length of query: 224
length of database: 9,014,727
effective HSP length: 93
effective length of query: 131
effective length of database: 5,575,959
effective search space: 730450629
effective search space used: 730450629
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0010.5


