

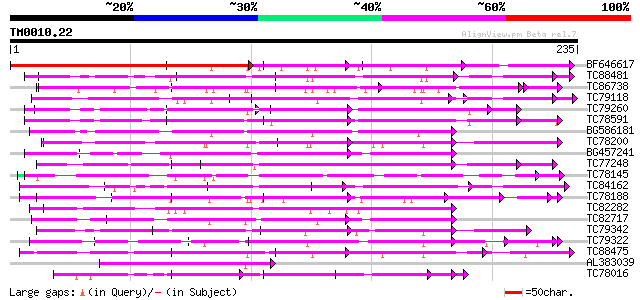
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0010.22
(235 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BF646617 similar to GP|20197268|gb| expressed protein {Arabidops... 196 5e-51
TC88481 similar to PIR|T00806|T02445 probable U4/U6 small nuclea... 87 6e-18
TC86738 similar to PIR|T00593|T02480 sec13-related protein At2g3... 80 7e-16
TC79118 similar to GP|16323188|gb|AAL15328.1 AT5g67320/K8K14_4 {... 73 1e-13
TC79260 weakly similar to GP|9759350|dbj|BAB10005.1 WD-repeat pr... 67 5e-12
TC78591 similar to GP|9759350|dbj|BAB10005.1 WD-repeat protein-l... 67 5e-12
BG586181 similar to GP|4689316|gb| peroxisomal targeting signal ... 66 1e-11
TC78200 similar to PIR|A84578|A84578 probable WD-40 repeat prote... 65 3e-11
BG457241 homologue to PIR|F84600|F8 coatomer alpha subunit [impo... 64 4e-11
TC77248 similar to GP|13489182|gb|AAK27816.1 putative WD-repeat ... 64 7e-11
TC78145 similar to GP|15294240|gb|AAK95297.1 AT3g18860/MCB22_3 {... 62 2e-10
TC84162 similar to GP|15810485|gb|AAL07130.1 putative WD40-repea... 61 4e-10
TC78188 similar to PIR|C84870|C84870 probable splicing factor [i... 60 6e-10
TC82282 similar to PIR|T08544|T08544 hypothetical protein F27B13... 60 9e-10
TC82717 weakly similar to GP|21539583|gb|AAM53344.1 putative coa... 57 5e-09
TC79342 similar to GP|10177096|dbj|BAB10430. Notchless protein h... 57 5e-09
TC79322 similar to GP|13605815|gb|AAK32893.1 At2g32700/F24L7.16 ... 56 1e-08
TC88475 similar to GP|20260442|gb|AAM13119.1 unknown protein {Ar... 56 1e-08
AL383039 weakly similar to PIR|A45442|A454 transport versicle fo... 55 2e-08
TC78016 similar to GP|21537191|gb|AAM61532.1 PRL1 protein {Arabi... 54 4e-08
>BF646617 similar to GP|20197268|gb| expressed protein {Arabidopsis
thaliana}, partial (23%)
Length = 533
Score = 196 bits (499), Expect = 5e-51
Identities = 91/101 (90%), Positives = 97/101 (95%)
Frame = +3
Query: 1 MERLELKEVQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWAC 60
MERLELKEVQ+LEGH DRVWSLDWNPATGHAG PLVFASCSGDKTVRIWEQ+LS+ L++C
Sbjct: 231 MERLELKEVQKLEGHTDRVWSLDWNPATGHAGIPLVFASCSGDKTVRIWEQNLSTNLFSC 410
Query: 61 KAVLDETHTRTVRSCAWSPSGKLLATASFDATTAIWENVGG 101
KA L+ETHTRTVRSCAWSPSGKLLATASFDATTAIWENVGG
Sbjct: 411 KATLEETHTRTVRSCAWSPSGKLLATASFDATTAIWENVGG 533
Score = 58.9 bits (141), Expect = 2e-09
Identities = 34/92 (36%), Positives = 51/92 (54%), Gaps = 8/92 (8%)
Frame = +3
Query: 106 VATLEGHENEVKSVSWN-ASG-----TLLATCSRDKSVWIWEMQ-PVNEFECVSVLQG-H 157
V LEGH + V S+ WN A+G + A+CS DK+V IWE N F C + L+ H
Sbjct: 255 VQKLEGHTDRVWSLDWNPATGHAGIPLVFASCSGDKTVRIWEQNLSTNLFSCKATLEETH 434
Query: 158 TQDVKMVKWHPTEDILISCSYDNSIKVWADEG 189
T+ V+ W P+ +L + S+D + +W + G
Sbjct: 435 TRTVRSCAWSPSGKLLATASFDATTAIWENVG 530
Score = 53.1 bits (126), Expect = 9e-08
Identities = 36/85 (42%), Positives = 46/85 (53%), Gaps = 9/85 (10%)
Frame = +3
Query: 66 ETHTRTVRSCAWSPSGK------LLATASFDATTAIWE-NVGGD-FECVATLEG-HENEV 116
E HT V S W+P+ + A+ S D T IWE N+ + F C ATLE H V
Sbjct: 267 EGHTDRVWSLDWNPATGHAGIPLVFASCSGDKTVRIWEQNLSTNLFSCKATLEETHTRTV 446
Query: 117 KSVSWNASGTLLATCSRDKSVWIWE 141
+S +W+ SG LLAT S D + IWE
Sbjct: 447 RSCAWSPSGKLLATASFDATTAIWE 521
Score = 44.7 bits (104), Expect = 3e-05
Identities = 26/94 (27%), Positives = 46/94 (48%), Gaps = 6/94 (6%)
Frame = +3
Query: 147 EFECVSVLQGHTQDVKMVKWHPTED------ILISCSYDNSIKVWADEGDSDDWQCVQTL 200
E + V L+GHT V + W+P + SCS D ++++W ++ + C TL
Sbjct: 243 ELKEVQKLEGHTDRVWSLDWNPATGHAGIPLVFASCSGDKTVRIWEQNLSTNLFSCKATL 422
Query: 201 GQPNNGHTSTVWALSFNASGDKMVTCSSNLLMAV 234
+ HT TV + +++ SG + T S + A+
Sbjct: 423 EET---HTRTVRSCAWSPSGKLLATASFDATTAI 515
>TC88481 similar to PIR|T00806|T02445 probable U4/U6 small nuclear
ribonucleoprotein [imported] - Arabidopsis thaliana,
partial (60%)
Length = 1836
Score = 87.0 bits (214), Expect = 6e-18
Identities = 63/216 (29%), Positives = 108/216 (49%), Gaps = 1/216 (0%)
Frame = +1
Query: 13 EGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETHTRTV 72
+GH +R+ + ++P+ + GT S DKT R+W+ + L +L E H+R+V
Sbjct: 1192 KGHLERLARIAFHPSGKYLGT------ASYDKTWRLWDVETEEEL-----LLQEGHSRSV 1338
Query: 73 RSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGTLLATCS 132
+ G L A+ DA +W+ G V LEGH + +S++ +G LAT
Sbjct: 1339 YGLDFHHDGSLAASCGLDALARVWDLRTG--RSVLALEGHVKSILGISFSPNGYHLATGG 1512
Query: 133 RDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTED-ILISCSYDNSIKVWADEGDS 191
D + IW+++ + + + + H+ + VK+ P E L++ SYD + KVW+ S
Sbjct: 1513 EDNTCRIWDLR---KKKSLYTIPAHSNLISQVKFEPQEGYFLVTASYDMTAKVWS----S 1671
Query: 192 DDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVTCS 227
D++ V+TL +GH + V +L G +VT S
Sbjct: 1672 RDFKPVKTL----SGHEAKVTSLDVLGDGGYIVTVS 1767
Score = 68.2 bits (165), Expect = 3e-12
Identities = 53/182 (29%), Positives = 90/182 (49%), Gaps = 2/182 (1%)
Frame = +1
Query: 7 KEVQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLD- 65
+E+ EGH+ V+ LD++ H G+ + ASC D R+W DL +G ++VL
Sbjct: 1300 EELLLQEGHSRSVYGLDFH----HDGS--LAASCGLDALARVW--DLRTG----RSVLAL 1443
Query: 66 ETHTRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNAS- 124
E H +++ ++SP+G LAT D T IW+ + + T+ H N + V +
Sbjct: 1444 EGHVKSILGISFSPNGYHLATGGEDNTCRIWDL--RKKKSLYTIPAHSNLISQVKFEPQE 1617
Query: 125 GTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKV 184
G L T S D + +W + +F+ V L GH V + +++ S+D +IK+
Sbjct: 1618 GYFLVTASYDMTAKVWSSR---DFKPVKTLSGHEAKVTSLDVLGDGGYIVTVSHDRTIKL 1788
Query: 185 WA 186
W+
Sbjct: 1789 WS 1794
Score = 65.9 bits (159), Expect = 1e-11
Identities = 44/166 (26%), Positives = 80/166 (47%), Gaps = 1/166 (0%)
Frame = +1
Query: 70 RTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWN-ASGTLL 128
R + C++S GK LAT SF T +W + + V+TL+GH V+++ L
Sbjct: 952 RPLTGCSFSRDGKGLATCSFTGATKLWSM--PNVKKVSTLKGHTQRATDVAYSPVHKNHL 1125
Query: 129 ATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVWADE 188
AT S D++ W Q + +GH + + + +HP+ L + SYD + ++W +
Sbjct: 1126 ATASADRTAKYWNDQGA----LLGTFKGHLERLARIAFHPSGKYLGTASYDKTWRLW--D 1287
Query: 189 GDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVTCSSNLLMAV 234
++++ +Q GH+ +V+ L F+ G +C + L V
Sbjct: 1288 VETEEELLLQ------EGHSRSVYGLDFHHDGSLAASCGLDALARV 1407
Score = 47.0 bits (110), Expect = 6e-06
Identities = 35/118 (29%), Positives = 54/118 (45%), Gaps = 1/118 (0%)
Frame = +1
Query: 111 GHENEVKSVSWNASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPT- 169
G + + S++ G LATCS + +W M V + VS L+GHTQ V + P
Sbjct: 943 GDDRPLTGCSFSRDGKGLATCSFTGATKLWSMPNVKK---VSTLKGHTQRATDVAYSPVH 1113
Query: 170 EDILISCSYDNSIKVWADEGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVTCS 227
++ L + S D + K W D+G L GH + ++F+ SG + T S
Sbjct: 1114 KNHLATASADRTAKYWNDQG---------ALLGTFKGHLERLARIAFHPSGKYLGTAS 1260
Score = 26.9 bits (58), Expect = 6.8
Identities = 15/45 (33%), Positives = 23/45 (50%)
Frame = +1
Query: 5 ELKEVQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIW 49
+ K V+ L GH +V SLD G G + S D+T+++W
Sbjct: 1675 DFKPVKTLSGHEAKVTSLD---VLGDGG---YIVTVSHDRTIKLW 1791
>TC86738 similar to PIR|T00593|T02480 sec13-related protein At2g30050 -
Arabidopsis thaliana, complete
Length = 1278
Score = 80.1 bits (196), Expect = 7e-16
Identities = 66/240 (27%), Positives = 103/240 (42%), Gaps = 22/240 (9%)
Frame = +2
Query: 12 LEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETHTRT 71
L GH VW + W H + ASCS D V +W++ + W V DE H +
Sbjct: 272 LTGHQGPVWEVAW----AHPKFGSLLASCSYDGRVILWKEG-NQNEWIQAHVFDE-HKSS 433
Query: 72 VRSCAWSPS--GKLLATASFDATTAIW-ENVGGDFECVATLEGHENEVKSVSW---NASG 125
V S AW+P G LA AS D +++ G ++ + H V SVSW A G
Sbjct: 434 VNSVAWAPHELGLCLACASSDGNISVFTARADGGWDTSRIDQAHPVGVTSVSWAPSMAPG 613
Query: 126 TLLAT---------CSR--DKSVWIWEMQPVN-EFECVSVLQGHTQDVKMVKWHPT---- 169
L+ + CS D +V +W++ + +C LQ H V+ V W P
Sbjct: 614 ALVGSGLLDPVQKLCSGGCDNTVKVWKLSNGQWKMDCFPALQKHNDWVRDVAWAPNLGLP 793
Query: 170 EDILISCSYDNSIKVWADEGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVTCSSN 229
+ + S S D + +W + D W+ G+ N + VW +S++ +G+ + N
Sbjct: 794 KSTIASASQDGKVIIWTAGKEGDHWE-----GKDLNDFKTPVWRVSWSLTGNILAVADGN 958
Score = 62.0 bits (149), Expect = 2e-10
Identities = 50/152 (32%), Positives = 67/152 (43%), Gaps = 4/152 (2%)
Frame = +2
Query: 68 HTRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNAS--G 125
H + + GK LATAS D T I + +ATL GH+ V V+W G
Sbjct: 149 HQDIIHDVSMDYYGKRLATASSDHTIKIIGVSNSASQHLATLTGHQGPVWEVAWAHPKFG 328
Query: 126 TLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTE-DILISC-SYDNSIK 183
+LLA+CS D V +W+ NE+ V H V V W P E + ++C S D +I
Sbjct: 329 SLLASCSYDGRVILWKEGNQNEWIQAHVFDEHKSSVNSVAWAPHELGLCLACASSDGNIS 508
Query: 184 VWADEGDSDDWQCVQTLGQPNNGHTSTVWALS 215
V+ D W + G TS WA S
Sbjct: 509 VFTARADG-GWDTSRIDQAHPVGVTSVSWAPS 601
Score = 53.9 bits (128), Expect = 5e-08
Identities = 47/158 (29%), Positives = 72/158 (44%), Gaps = 15/158 (9%)
Frame = +2
Query: 13 EGHNDRVWSLDWNPA------TGHAGTPLVFASCSG--DKTVRIWEQDLSSGLWA--CKA 62
+ H V S+ W P+ G V CSG D TV++W+ LS+G W C
Sbjct: 557 QAHPVGVTSVSWAPSMAPGALVGSGLLDPVQKLCSGGCDNTVKVWK--LSNGQWKMDCFP 730
Query: 63 VLDETHTRTVRSCAWSPSGKL----LATASFDATTAIWE-NVGGDFECVATLEGHENEVK 117
L + H VR AW+P+ L +A+AS D IW GD L + V
Sbjct: 731 ALQK-HNDWVRDVAWAPNLGLPKSTIASASQDGKVIIWTAGKEGDHWEGKDLNDFKTPVW 907
Query: 118 SVSWNASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQ 155
VSW+ +G +LA + +V +W+ E++ V+ ++
Sbjct: 908 RVSWSLTGNILAVADGNNNVTLWKEAVDGEWQQVTTVE 1021
Score = 48.1 bits (113), Expect = 3e-06
Identities = 32/105 (30%), Positives = 51/105 (48%), Gaps = 2/105 (1%)
Frame = +2
Query: 111 GHENEVKSVSWNASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKW-HPT 169
GH++ + VS + G LAT S D ++ I + + + ++ L GH V V W HP
Sbjct: 146 GHQDIIHDVSMDYYGKRLATASSDHTIKIIGVSN-SASQHLATLTGHQGPVWEVAWAHPK 322
Query: 170 -EDILISCSYDNSIKVWADEGDSDDWQCVQTLGQPNNGHTSTVWA 213
+L SCSYD + +W EG+ ++W + + S WA
Sbjct: 323 FGSLLASCSYDGRVILW-KEGNQNEWIQAHVFDEHKSSVNSVAWA 454
>TC79118 similar to GP|16323188|gb|AAL15328.1 AT5g67320/K8K14_4 {Arabidopsis
thaliana}, partial (44%)
Length = 1098
Score = 72.8 bits (177), Expect = 1e-13
Identities = 53/184 (28%), Positives = 89/184 (47%), Gaps = 9/184 (4%)
Frame = +2
Query: 10 QRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETHT 69
Q GH V + W+P T + ASCS D T +IW L + H+
Sbjct: 299 QTFAGHQGEVNCVKWDP------TGTLLASCSDDITAKIWSVKQDKYLHDFRE-----HS 445
Query: 70 RTVRSCAWSPSGK---------LLATASFDATTAIWENVGGDFECVATLEGHENEVKSVS 120
+ + + WSP+G LLA+ASFD+T +W+ G + + +L GH + V SV+
Sbjct: 446 KEIYTIRWSPTGPGTNNPNKKLLLASASFDSTVKLWDIELG--KLIHSLNGHRHPVYSVA 619
Query: 121 WNASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDN 180
++ +G +A+ S DKS+ IW ++ E + + G + + V W+ D + +C +N
Sbjct: 620 FSPNGEYIASGSLDKSLHIWSLK---EGKIIRTYNG-SGGIFEVCWNKEGDKIAACFANN 787
Query: 181 SIKV 184
+ V
Sbjct: 788 IVCV 799
Score = 66.2 bits (160), Expect = 1e-11
Identities = 38/136 (27%), Positives = 74/136 (53%), Gaps = 9/136 (6%)
Frame = +2
Query: 101 GDFECVATLEGHENEVKSVSWNASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQD 160
G+ T GH+ EV V W+ +GTLLA+CS D + IW V + + + + H+++
Sbjct: 281 GENHPTQTFAGHQGEVNCVKWDPTGTLLASCSDDITAKIWS---VKQDKYLHDFREHSKE 451
Query: 161 VKMVKWHPT---------EDILISCSYDNSIKVWADEGDSDDWQCVQTLGQPNNGHTSTV 211
+ ++W PT + +L S S+D+++K+W D + + + +L NGH V
Sbjct: 452 IYTIRWSPTGPGTNNPNKKLLLASASFDSTVKLW----DIELGKLIHSL----NGHRHPV 607
Query: 212 WALSFNASGDKMVTCS 227
++++F+ +G+ + + S
Sbjct: 608 YSVAFSPNGEYIASGS 655
Score = 65.9 bits (159), Expect = 1e-11
Identities = 62/243 (25%), Positives = 107/243 (43%), Gaps = 17/243 (6%)
Frame = +2
Query: 10 QRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETH- 68
Q+ H+ +DW T FAS S D + + CK + E H
Sbjct: 176 QQFGFHSGPTLDVDWRTNTS-------FASSSTDNMI-----------YVCK--IGENHP 295
Query: 69 TRT-------VRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSW 121
T+T V W P+G LLA+ S D T IW +V D + + H E+ ++ W
Sbjct: 296 TQTFAGHQGEVNCVKWDPTGTLLASCSDDITAKIW-SVKQD-KYLHDFREHSKEIYTIRW 469
Query: 122 NASGT---------LLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDI 172
+ +G LLA+ S D +V +W+++ + + L GH V V + P +
Sbjct: 470 SPTGPGTNNPNKKLLLASASFDSTVKLWDIELG---KLIHSLNGHRHPVYSVAFSPNGEY 640
Query: 173 LISCSYDNSIKVWADEGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVTCSSNLLM 232
+ S S D S+ +W+ + + ++T NG + ++ + +N GDK+ C +N ++
Sbjct: 641 IASGSLDKSLHIWS----LKEGKIIRTY----NG-SGGIFEVCWNKEGDKIAACFANNIV 793
Query: 233 AVM 235
V+
Sbjct: 794 CVL 802
Score = 47.8 bits (112), Expect = 4e-06
Identities = 30/99 (30%), Positives = 46/99 (46%)
Frame = +2
Query: 92 TTAIWENVGGDFECVATLEGHENEVKSVSWNASGTLLATCSRDKSVWIWEMQPVNEFECV 151
+TAI +V + EC H V W + T A+ S D +++ + + E
Sbjct: 134 STAIVWDVKAE-ECKQQFGFHSGPTLDVDWRTN-TSFASSSTDNMIYVCK---IGENHPT 298
Query: 152 SVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVWADEGD 190
GH +V VKW PT +L SCS D + K+W+ + D
Sbjct: 299 QTFAGHQGEVNCVKWDPTGTLLASCSDDITAKIWSVKQD 415
>TC79260 weakly similar to GP|9759350|dbj|BAB10005.1 WD-repeat protein-like
{Arabidopsis thaliana}, partial (50%)
Length = 1227
Score = 67.4 bits (163), Expect = 5e-12
Identities = 43/137 (31%), Positives = 67/137 (48%), Gaps = 2/137 (1%)
Frame = +2
Query: 66 ETHTRTVRSCAWSPSGKLLATASFDATTAIWE-NVGGDFECVATLEGHENEVKSVSWNAS 124
+ H V +S GK LATAS D T IWE + L GH+ V SVSW+ +
Sbjct: 536 DAHDDEVWFVQFSHDGKYLATASNDRTAIIWEVDTNDGLSMKHKLSGHQKSVSSVSWSPN 715
Query: 125 GTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKV 184
G L TC +++V W+ V+ +C+ V + + + W P ++S D SI +
Sbjct: 716 GQELLTCGVEEAVRRWD---VSTGKCLQVYEKNGSGLISCAWFPCGKYILSGLSDKSICM 886
Query: 185 WADEG-DSDDWQCVQTL 200
W +G + + W+ +TL
Sbjct: 887 WELDGKEVESWKGQKTL 937
Score = 65.1 bits (157), Expect = 2e-11
Identities = 42/136 (30%), Positives = 71/136 (51%)
Frame = +2
Query: 7 KEVQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDE 66
+ +Q L+ H+D VW + ++ H G L A+ S D+T IWE D + GL + K L
Sbjct: 518 RTLQILDAHDDEVWFVQFS----HDGKYL--ATASNDRTAIIWEVDTNDGL-SMKHKL-S 673
Query: 67 THTRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGT 126
H ++V S +WSP+G+ L T + W+ G +C+ E + + + S +W G
Sbjct: 674 GHQKSVSSVSWSPNGQELLTCGVEEAVRRWDVSTG--KCLQVYEKNGSGLISCAWFPCGK 847
Query: 127 LLATCSRDKSVWIWEM 142
+ + DKS+ +WE+
Sbjct: 848 YILSGLSDKSICMWEL 895
Score = 60.5 bits (145), Expect = 6e-10
Identities = 30/104 (28%), Positives = 50/104 (47%)
Frame = +2
Query: 109 LEGHENEVKSVSWNASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHP 168
L+ H++EV V ++ G LAT S D++ IWE+ + L GH + V V W P
Sbjct: 533 LDAHDDEVWFVQFSHDGKYLATASNDRTAIIWEVDTNDGLSMKHKLSGHQKSVSSVSWSP 712
Query: 169 TEDILISCSYDNSIKVWADEGDSDDWQCVQTLGQPNNGHTSTVW 212
L++C + +++ W D +C+Q + +G S W
Sbjct: 713 NGQELLTCGVEEAVRRW----DVSTGKCLQVYEKNGSGLISCAW 832
Score = 45.1 bits (105), Expect = 2e-05
Identities = 30/94 (31%), Positives = 48/94 (50%)
Frame = +2
Query: 11 RLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETHTR 70
+L GH V S+ W+P G L+ +C ++ VR W D+S+G C V ++ +
Sbjct: 665 KLSGHQKSVSSVSWSPN----GQELL--TCGVEEAVRRW--DVSTG--KCLQVYEKNGSG 814
Query: 71 TVRSCAWSPSGKLLATASFDATTAIWENVGGDFE 104
+ SCAW P GK + + D + +WE G + E
Sbjct: 815 LI-SCAWFPCGKYILSGLSDKSICMWELDGKEVE 913
Score = 31.2 bits (69), Expect = 0.36
Identities = 18/76 (23%), Positives = 40/76 (51%)
Frame = +2
Query: 151 VSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVWADEGDSDDWQCVQTLGQPNNGHTST 210
+ +L H +V V++ L + S D + +W E D++D ++ +GH +
Sbjct: 524 LQILDAHDDEVWFVQFSHDGKYLATASNDRTAIIW--EVDTNDGLSMK---HKLSGHQKS 688
Query: 211 VWALSFNASGDKMVTC 226
V ++S++ +G +++TC
Sbjct: 689 VSSVSWSPNGQELLTC 736
>TC78591 similar to GP|9759350|dbj|BAB10005.1 WD-repeat protein-like
{Arabidopsis thaliana}, partial (17%)
Length = 916
Score = 67.4 bits (163), Expect = 5e-12
Identities = 49/167 (29%), Positives = 83/167 (49%), Gaps = 3/167 (1%)
Frame = +2
Query: 66 ETHTRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVA-TLEGHENEVKSVSWNAS 124
ETH V +S +GK LA+AS D T IWE VG + CV L GH+ + SVSW+ +
Sbjct: 455 ETHDDEVWFVQFSHNGKYLASASNDQTAIIWE-VGVNGVCVKHRLSGHQKPISSVSWSPN 631
Query: 125 GTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKV 184
L TC ++S+ W+ V+ +C+ + + + W P+ ++S D SI +
Sbjct: 632 DEELLTCGVEESIRRWD---VSTGKCLQIYEKAGAGLVSCTWFPSGKHILSGLSDKSICM 802
Query: 185 WADEG-DSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVT-CSSN 229
W +G + + W+ ++L + L A G+++++ C N
Sbjct: 803 WELDGKEVESWKGQKSL---------KISDLDITADGEEIISICKPN 916
Score = 59.7 bits (143), Expect = 9e-10
Identities = 34/107 (31%), Positives = 54/107 (49%)
Frame = +2
Query: 106 VATLEGHENEVKSVSWNASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVK 165
V LE H++EV V ++ +G LA+ S D++ IWE+ VN L GH + + V
Sbjct: 443 VQILETHDDEVWFVQFSHNGKYLASASNDQTAIIWEVG-VNGVCVKHRLSGHQKPISSVS 619
Query: 166 WHPTEDILISCSYDNSIKVWADEGDSDDWQCVQTLGQPNNGHTSTVW 212
W P ++ L++C + SI+ W D +C+Q + G S W
Sbjct: 620 WSPNDEELLTCGVEESIRRW----DVSTGKCLQIYEKAGAGLVSCTW 748
Score = 57.4 bits (137), Expect = 5e-09
Identities = 40/136 (29%), Positives = 65/136 (47%)
Frame = +2
Query: 7 KEVQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDE 66
+ VQ LE H+D VW + ++ H G L AS S D+T IWE ++ C
Sbjct: 437 RTVQILETHDDEVWFVQFS----HNGKYL--ASASNDQTAIIWEVGVNG---VCVKHRLS 589
Query: 67 THTRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGT 126
H + + S +WSP+ + L T + + W+ G +C+ E + S +W SG
Sbjct: 590 GHQKPISSVSWSPNDEELLTCGVEESIRRWDVSTG--KCLQIYEKAGAGLVSCTWFPSGK 763
Query: 127 LLATCSRDKSVWIWEM 142
+ + DKS+ +WE+
Sbjct: 764 HILSGLSDKSICMWEL 811
>BG586181 similar to GP|4689316|gb| peroxisomal targeting signal type 2
receptor {Arabidopsis thaliana}, partial (56%)
Length = 804
Score = 65.9 bits (159), Expect = 1e-11
Identities = 58/180 (32%), Positives = 82/180 (45%), Gaps = 3/180 (1%)
Frame = +1
Query: 9 VQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETH 68
V+ + H V+S WNP HA VFAS SGD TVRIW+ A H
Sbjct: 16 VRTFKEHAYCVYSSVWNPR--HAD---VFASASGDCTVRIWDVREPGSTMILPA-----H 165
Query: 69 TRTVRSCAWSPSGK-LLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNAS-GT 126
+ SC W+ + ++AT S D + +W+ V VA L GH V+ V ++
Sbjct: 166 EHEILSCDWNKYDECIIATGSVDKSVKVWD-VRSYRVPVAVLNGHGYAVRKVKFSPHVRN 342
Query: 127 LLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHP-TEDILISCSYDNSIKVW 185
LL +CS D +V +W+ + E VS HT+ V E +L S +D + VW
Sbjct: 343 LLVSCSYDMTVCVWDF--MVEDALVSRYDHHTEFAVGVDMSVLVEGLLASTGWDELVYVW 516
>TC78200 similar to PIR|A84578|A84578 probable WD-40 repeat protein
[imported] - Arabidopsis thaliana, partial (80%)
Length = 1824
Score = 64.7 bits (156), Expect = 3e-11
Identities = 47/175 (26%), Positives = 85/175 (47%), Gaps = 4/175 (2%)
Frame = +1
Query: 15 HNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETHTRTVRS 74
H D +++DW+P LV C + ++ +WE S+ W + HT +V
Sbjct: 703 HKDEGYAIDWSPLVPGR---LVSGDC--NNSIYLWE-PTSAATWNIEKTPFTGHTDSVED 864
Query: 75 CAWSPS-GKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWN-ASGTLLATCS 132
WSP+ + A+ S D + AIW+ G A+ + H+ +V +SWN + +LA+ S
Sbjct: 865 LQWSPTEPHVFASCSVDKSIAIWDTRLGR-SPAASFQAHKADVNVLSWNRLASCMLASGS 1041
Query: 133 RDKSVWIWEMQPVNEFE-CVSVLQGHTQDVKMVKWHPTE-DILISCSYDNSIKVW 185
D ++ I +++ + E + V+ + H + ++W P E L S DN + +W
Sbjct: 1042DDGTISIRDLRLLKEGDSVVAHFEYHKHPITSIEWSPHEASSLAVSSADNQLTIW 1206
Score = 58.9 bits (141), Expect = 2e-09
Identities = 43/134 (32%), Positives = 64/134 (47%), Gaps = 5/134 (3%)
Frame = +1
Query: 14 GHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETHTRTVR 73
GH D V L W+P H VFASCS DK++ IW+ L A + H V
Sbjct: 841 GHTDSVEDLQWSPTEPH-----VFASCSVDKSIAIWDTRLGRS----PAASFQAHKADVN 993
Query: 74 SCAWSP-SGKLLATASFDATTAIWE---NVGGDFECVATLEGHENEVKSVSWNA-SGTLL 128
+W+ + +LA+ S D T +I + GD VA E H++ + S+ W+ + L
Sbjct: 994 VLSWNRLASCMLASGSDDGTISIRDLRLLKEGD-SVVAHFEYHKHPITSIEWSPHEASSL 1170
Query: 129 ATCSRDKSVWIWEM 142
A S D + IW++
Sbjct: 1171AVSSADNQLTIWDL 1212
Score = 42.0 bits (97), Expect = 2e-04
Identities = 31/122 (25%), Positives = 55/122 (44%), Gaps = 4/122 (3%)
Frame = +1
Query: 112 HENEVKSVSWN--ASGTLLATCSRDKSVWIWEMQPVNEFECVSV-LQGHTQDVKMVKWHP 168
H++E ++ W+ G L++ + S+++WE + GHT V+ ++W P
Sbjct: 703 HKDEGYAIDWSPLVPGRLVSG-DCNNSIYLWEPTSAATWNIEKTPFTGHTDSVEDLQWSP 879
Query: 169 TE-DILISCSYDNSIKVWADEGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVTCS 227
TE + SCS D SI +W D + ++ H + V LS+N M+
Sbjct: 880 TEPHVFASCSVDKSIAIW-------DTRLGRSPAASFQAHKADVNVLSWNRLASCMLASG 1038
Query: 228 SN 229
S+
Sbjct: 1039SD 1044
>BG457241 homologue to PIR|F84600|F8 coatomer alpha subunit [imported] -
Arabidopsis thaliana, partial (15%)
Length = 654
Score = 64.3 bits (155), Expect = 4e-11
Identities = 44/181 (24%), Positives = 84/181 (46%), Gaps = 2/181 (1%)
Frame = +1
Query: 7 KEVQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLD- 65
K + + E ++RV L ++P P + AS +++W+ + + ++D
Sbjct: 70 KMLTKFETKSNRVKGLSFHPKR-----PWILASLHSG-VIQLWDYRMGT-------LIDK 210
Query: 66 -ETHTRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNAS 124
+ H VR + S L + D +W C+ TL GH + +++V ++
Sbjct: 211 FDEHDGPVRGVHFHNSQPLFVSGGDDYKIKVWNYKL--HRCLFTLLGHLDYIRTVQFHHE 384
Query: 125 GTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKV 184
+ + S D+++ IW Q C+SVL GH V +HP +D+++S S D +++V
Sbjct: 385 SPWIVSASDDQTIRIWNWQSRT---CISVLTGHNHYVMCALFHPKDDLVVSASLDQTVRV 555
Query: 185 W 185
W
Sbjct: 556 W 558
Score = 42.4 bits (98), Expect = 2e-04
Identities = 32/113 (28%), Positives = 52/113 (45%)
Frame = +1
Query: 30 HAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETHTRTVRSCAWSPSGKLLATASF 89
H PL F S D +++W L L+ LD + RTV+ SP + +AS
Sbjct: 250 HNSQPL-FVSGGDDYKIKVWNYKLHRCLFTLLGHLD--YIRTVQFHHESP---WIVSASD 411
Query: 90 DATTAIWENVGGDFECVATLEGHENEVKSVSWNASGTLLATCSRDKSVWIWEM 142
D T IW C++ L GH + V ++ L+ + S D++V +W++
Sbjct: 412 DQTIRIWN--WQSRTCISVLTGHNHYVMCALFHPKDDLVVSASLDQTVRVWDI 564
>TC77248 similar to GP|13489182|gb|AAK27816.1 putative WD-repeat containing
protein {Oryza sativa}, partial (92%)
Length = 2005
Score = 63.5 bits (153), Expect = 7e-11
Identities = 36/133 (27%), Positives = 64/133 (48%)
Frame = +1
Query: 80 SGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGTLLATCSRDKSVWI 139
S L+AT + D I++ G + +ATL GH +V SV + G + T S DK+V +
Sbjct: 793 SKDLIATGNIDTNAVIFDRPSG--QVLATLTGHSKKVTSVKFVGQGESIITGSADKTVRL 966
Query: 140 WEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVWADEGDSDDWQCVQT 199
W+ + C +L+ H+ +V+ V H T + ++ S D S + + C+
Sbjct: 967 WQGSDDGHYNCKQILKDHSAEVEAVTVHATNNYFVTASLDGSWCFY----ELSSGTCLTQ 1134
Query: 200 LGQPNNGHTSTVW 212
+ + G+TS +
Sbjct: 1135VSDSSEGYTSAAF 1173
Score = 56.6 bits (135), Expect = 8e-09
Identities = 38/161 (23%), Positives = 75/161 (45%), Gaps = 1/161 (0%)
Frame = +1
Query: 68 HTRTVRSCAWSPSGKLLATASFDATTAIWENVG-GDFECVATLEGHENEVKSVSWNASGT 126
H++ V S + G+ + T S D T +W+ G + C L+ H EV++V+ +A+
Sbjct: 883 HSKKVTSVKFVGQGESIITGSADKTVRLWQGSDDGHYNCKQILKDHSAEVEAVTVHATNN 1062
Query: 127 LLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVWA 186
T S D S W ++ C++ + ++ +HP IL + + D+ +K+W
Sbjct: 1063 YFVTASLDGS---WCFYELSSGTCLTQVSDSSEGYTSAAFHPDGLILGTGTTDSLVKIWD 1233
Query: 187 DEGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVTCS 227
+ ++ + +GH V A+SF+ +G + T +
Sbjct: 1234 VKSQANVAKF--------DGHVGHVTAISFSENGYYLATAA 1332
Score = 53.5 bits (127), Expect = 7e-08
Identities = 44/174 (25%), Positives = 74/174 (42%)
Frame = +1
Query: 12 LEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETHTRT 71
L GH+ +V S+ + G + S DKTVR+W Q G + CK +L + H+
Sbjct: 874 LTGHSKKVTSVKF------VGQGESIITGSADKTVRLW-QGSDDGHYNCKQILKD-HSAE 1029
Query: 72 VRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGTLLATC 131
V + + TAS D + +E G C+ + S +++ G +L T
Sbjct: 1030 VEAVTVHATNNYFVTASLDGSWCFYELSSG--TCLTQVSDSSEGYTSAAFHPDGLILGTG 1203
Query: 132 SRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVW 185
+ D V IW+ V V+ GH V + + L + ++D +K+W
Sbjct: 1204 TTDSLVKIWD---VKSQANVAKFDGHVGHVTAISFSENGYYLATAAHD-GVKLW 1353
>TC78145 similar to GP|15294240|gb|AAK95297.1 AT3g18860/MCB22_3 {Arabidopsis
thaliana}, partial (93%)
Length = 2633
Score = 61.6 bits (148), Expect = 2e-10
Identities = 60/224 (26%), Positives = 94/224 (41%)
Frame = +3
Query: 7 KEVQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDE 66
++VQ L+GH +V TG S S D T+R W W E
Sbjct: 390 EKVQSLKGHQLQV--------TGIVVDDGDLVSSSMDCTLRRWRNGQCIETW-------E 524
Query: 67 THTRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGT 126
H +++ P+G+L+ T S D T W+ C+ T +GH + V+ ++ +
Sbjct: 525 AHKGPIQAVIKLPTGELV-TGSSDTTLKTWKGK----TCLHTFQGHADTVRGLAVMSDLG 689
Query: 127 LLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVWA 186
+L+ S D S+ +W + E V GHT V V H + +++S S D K+W
Sbjct: 690 ILSA-SHDGSLRLWAVSGEGLMEMV----GHTAIVYSVDSHAS-GLIVSGSEDRFAKIWK 851
Query: 187 DEGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVTCSSNL 230
D CVQ++ P VW F +GD + CS +
Sbjct: 852 DG------VCVQSIEHPG-----CVWDTKFMENGDIVTACSDGI 950
Score = 53.5 bits (127), Expect = 7e-08
Identities = 68/268 (25%), Positives = 107/268 (39%), Gaps = 51/268 (19%)
Frame = +3
Query: 4 LELKEVQ---RLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWAC 60
++ KE Q L GH D V + T G A+ S DKT+R+W +D
Sbjct: 96 IDFKEYQLRCELHGHEDDVRGI-----TICGGDNGGIATSSRDKTIRLWTRDNR------ 242
Query: 61 KAVLDET---HTRTVRSCAWSP-----------SGKL----------------------- 83
K VLD+ HT V AW P SG +
Sbjct: 243 KFVLDKVLVGHTSFVGPIAWIPPNPDLPHGGVVSGGMDTLVLVWDLNSGEKVQSLKGHQL 422
Query: 84 -----------LATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGTLLATCS 132
L ++S D T W N +C+ T E H+ +++V +G L+ T S
Sbjct: 423 QVTGIVVDDGDLVSSSMDCTLRRWRN----GQCIETWEAHKGPIQAVIKLPTGELV-TGS 587
Query: 133 RDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVWADEGDSD 192
D ++ W+ + C+ QGH V+ + IL S S+D S+++WA G+
Sbjct: 588 SDTTLKTWKGK-----TCLHTFQGHADTVRGLAVMSDLGIL-SASHDGSLRLWAVSGEG- 746
Query: 193 DWQCVQTLGQPNNGHTSTVWALSFNASG 220
++ + GHT+ V+++ +ASG
Sbjct: 747 ---LMEMV-----GHTAIVYSVDSHASG 806
>TC84162 similar to GP|15810485|gb|AAL07130.1 putative WD40-repeat protein
{Arabidopsis thaliana}, partial (29%)
Length = 628
Score = 60.8 bits (146), Expect = 4e-10
Identities = 37/150 (24%), Positives = 68/150 (44%)
Frame = +1
Query: 83 LLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGTLLATCSRDKSVWIWEM 142
L+ + S D T +W D V +GH+ + SV ++ + T S DK++ IW
Sbjct: 28 LVCSGSQDRTACVWRLP--DLVSVVVFKGHKRGIWSVEFSPVDQCVVTASGDKTIRIWA- 198
Query: 143 QPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVWADEGDSDDWQCVQTLGQ 202
+++ C+ +GHT V + ++SC D +K+W + + +CV T
Sbjct: 199 --ISDGSCLKTFEGHTSSVLRALFVTRGTQIVSCGADGLVKLWTVKSN----ECVATY-- 354
Query: 203 PNNGHTSTVWALSFNASGDKMVTCSSNLLM 232
+ H VWAL+ + + T S+ ++
Sbjct: 355 --DNHEDKVWALAVGRKTETLATGGSDAVV 438
Score = 60.5 bits (145), Expect = 6e-10
Identities = 46/156 (29%), Positives = 67/156 (42%), Gaps = 3/156 (1%)
Frame = +1
Query: 40 CSG--DKTVRIWE-QDLSSGLWACKAVLDETHTRTVRSCAWSPSGKLLATASFDATTAIW 96
CSG D+T +W DL S V+ + H R + S +SP + + TAS D T IW
Sbjct: 34 CSGSQDRTACVWRLPDLVS------VVVFKGHKRGIWSVEFSPVDQCVVTASGDKTIRIW 195
Query: 97 ENVGGDFECVATLEGHENEVKSVSWNASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQG 156
G C+ T EGH + V + GT + +C D V +W V ECV+
Sbjct: 196 AISDGS--CLKTFEGHTSSVLRALFVTRGTQIVSCGADGLVKLWT---VKSNECVATYDN 360
Query: 157 HTQDVKMVKWHPTEDILISCSYDNSIKVWADEGDSD 192
H V + + L + D + +W D +D
Sbjct: 361 HEDKVWALAVGRKTETLATGGSDAVVNLWLDSTAAD 468
Score = 55.1 bits (131), Expect = 2e-08
Identities = 40/136 (29%), Positives = 62/136 (45%)
Frame = +1
Query: 5 ELKEVQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVL 64
+L V +GH +WS++++P T SGDKT+RIW +S G +C
Sbjct: 79 DLVSVVVFKGHKRGIWSVEFSPVDQCVVT------ASGDKTIRIWA--ISDG--SCLKTF 228
Query: 65 DETHTRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNAS 124
E HT +V + G + + D +W ECVAT + HE++V +++
Sbjct: 229 -EGHTSSVLRALFVTRGTQIVSCGADGLVKLWTVKSN--ECVATYDNHEDKVWALAVGRK 399
Query: 125 GTLLATCSRDKSVWIW 140
LAT D V +W
Sbjct: 400 TETLATGGSDAVVNLW 447
Score = 51.2 bits (121), Expect = 3e-07
Identities = 28/107 (26%), Positives = 58/107 (54%)
Frame = +1
Query: 126 TLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVW 185
+L+ + S+D++ +W + + V V +GH + + V++ P + +++ S D +I++W
Sbjct: 25 SLVCSGSQDRTACVWRLP---DLVSVVVFKGHKRGIWSVEFSPVDQCVVTASGDKTIRIW 195
Query: 186 ADEGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVTCSSNLLM 232
A D C++T GHTS+V F G ++V+C ++ L+
Sbjct: 196 A----ISDGSCLKTF----EGHTSSVLRALFVTRGTQIVSCGADGLV 312
>TC78188 similar to PIR|C84870|C84870 probable splicing factor [imported] -
Arabidopsis thaliana, partial (95%)
Length = 1436
Score = 60.5 bits (145), Expect = 6e-10
Identities = 65/270 (24%), Positives = 109/270 (40%), Gaps = 45/270 (16%)
Frame = +1
Query: 5 ELKEVQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVL 64
+ K L+GH + V L W GT ++ S S DKT+R+W D +G K V
Sbjct: 409 DCKNFMVLKGHKNAVLDLHWTSD----GTQII--SASPDKTLRLW--DTETGKQIKKMV- 561
Query: 65 DETHTRTVRSCAWSPSG-KLLATASFDATTAIWE-------------------------- 97
H V SC + G L+ + S D T +W+
Sbjct: 562 --EHLSYVNSCCPTRRGPPLVVSGSDDGTAKLWDMRQRGSIQTFPDKYQITAVSFSDASD 735
Query: 98 -----NVGGDF--------ECVATLEGHENEVKSVSWNASGTLLATCSRDKSVWIWEMQP 144
+ D E TL+GH++ + S+ + G+ L T D + IW+M+P
Sbjct: 736 KIYTGGIDNDVKIWDLRKGEVTMTLQGHQDMITSMQLSPDGSYLLTNGMDCKLCIWDMRP 915
Query: 145 -VNEFECVSVLQGHTQDVK--MVK--WHPTEDILISCSYDNSIKVWADEGDSDDWQCVQT 199
+ CV +L+GH + + ++K W P + + S D + +W D+ + +
Sbjct: 916 YAPQNRCVKILEGHQHNFEKNLLKCGWSPDGSKVTAGSSDRMVYIW----DTTSRRILYK 1083
Query: 200 LGQPNNGHTSTVWALSFNASGDKMVTCSSN 229
L GH +V F+ + + +CSS+
Sbjct: 1084L----PGHNGSVNECVFHPNEPIVGSCSSD 1161
Score = 58.9 bits (141), Expect = 2e-09
Identities = 44/146 (30%), Positives = 58/146 (39%), Gaps = 6/146 (4%)
Frame = +1
Query: 43 DKTVRIWEQDLSSGLWACKAVLDETHTRTVRSCAWSPSGKLLATASFDATTAIWEN--VG 100
D V+IW DL G + H + S SP G L T D IW+
Sbjct: 757 DNDVKIW--DLRKGEVTMTL---QGHQDMITSMQLSPDGSYLLTNGMDCKLCIWDMRPYA 921
Query: 101 GDFECVATLEGH----ENEVKSVSWNASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQG 156
CV LEGH E + W+ G+ + S D+ V+IW+ + L G
Sbjct: 922 PQNRCVKILEGHQHNFEKNLLKCGWSPDGSKVTAGSSDRMVYIWD---TTSRRILYKLPG 1092
Query: 157 HTQDVKMVKWHPTEDILISCSYDNSI 182
H V +HP E I+ SCS D I
Sbjct: 1093HNGSVNECVFHPNEPIVGSCSSDKQI 1170
Score = 57.8 bits (138), Expect = 4e-09
Identities = 39/158 (24%), Positives = 75/158 (46%)
Frame = +1
Query: 68 HTRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGTL 127
H V + ++P+G ++A+ S D +W NV GD + L+GH+N V + W + GT
Sbjct: 310 HQSAVYTMKFNPTGSVVASGSHDKEIFLW-NVHGDCKNFMVLKGHKNAVLDLHWTSDGTQ 486
Query: 128 LATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVWAD 187
+ + S DK++ +W+ + + + + + + P +++S S D + K+W
Sbjct: 487 IISASPDKTLRLWDTETGKQIKKMVEHLSYVNSCCPTRRGP--PLVVSGSDDGTAKLW-- 654
Query: 188 EGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVT 225
D +QT + A+SF+ + DK+ T
Sbjct: 655 --DMRQRGSIQTFPDKYQ-----ITAVSFSDASDKIYT 747
Score = 49.7 bits (117), Expect = 1e-06
Identities = 26/100 (26%), Positives = 53/100 (53%)
Frame = +1
Query: 106 VATLEGHENEVKSVSWNASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVK 165
+ L GH++ V ++ +N +G+++A+ S DK +++W + + + VL+GH V +
Sbjct: 292 IMLLTGHQSAVYTMKFNPTGSVVASGSHDKEIFLWNVH--GDCKNFMVLKGHKNAVLDLH 465
Query: 166 WHPTEDILISCSYDNSIKVWADEGDSDDWQCVQTLGQPNN 205
W +IS S D ++++W E + V+ L N+
Sbjct: 466 WTSDGTQIISASPDKTLRLWDTETGKQIKKMVEHLSYVNS 585
Score = 47.0 bits (110), Expect = 6e-06
Identities = 30/134 (22%), Positives = 56/134 (41%), Gaps = 3/134 (2%)
Frame = +1
Query: 12 LEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLD---ETH 68
L+GH D + S+ +P + T + D + IW+ + C +L+
Sbjct: 808 LQGHQDMITSMQLSPDGSYLLTNGM------DCKLCIWDMRPYAPQNRCVKILEGHQHNF 969
Query: 69 TRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGTLL 128
+ + C WSP G + S D IW+ + L GH V ++ + ++
Sbjct: 970 EKNLLKCGWSPDGSKVTAGSSDRMVYIWDTTSR--RILYKLPGHNGSVNECVFHPNEPIV 1143
Query: 129 ATCSRDKSVWIWEM 142
+CS DK +++ E+
Sbjct: 1144GSCSSDKQIYLGEI 1185
>TC82282 similar to PIR|T08544|T08544 hypothetical protein F27B13.70 -
Arabidopsis thaliana, partial (84%)
Length = 904
Score = 59.7 bits (143), Expect = 9e-10
Identities = 55/185 (29%), Positives = 83/185 (44%), Gaps = 14/185 (7%)
Frame = +2
Query: 15 HNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDET---HTRT 71
H+D VW++ W PAT A P + + S D+TVR+W+ D VL+ T H
Sbjct: 113 HDDSVWAVTWAPAT--ATRPPLLLTGSLDETVRLWKSD--------DLVLERTNTGHCLG 262
Query: 72 VRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGTLLATC 131
V S A P G + A++S D+ +++ +ATLE +EV + ++ G +LA
Sbjct: 263 VASVAAHPLGSIAASSSLDSFVRVFDVDSN--ATIATLEAPPSEVWQMRFDPKGAILAVA 436
Query: 132 ---SRDKSVW---IWEMQ-----PVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDN 180
S ++W WE+ P E S G + V V W P L S D
Sbjct: 437 GGGSASVNLWDTSTWELVVTLSIPRVEGPKPSDKSGSKKFVLSVAWSPDGKRLACGSMDG 616
Query: 181 SIKVW 185
+I V+
Sbjct: 617 TISVF 631
Score = 44.3 bits (103), Expect = 4e-05
Identities = 46/190 (24%), Positives = 80/190 (41%), Gaps = 13/190 (6%)
Frame = +2
Query: 9 VQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVL---- 64
+ LE VW + ++P G L A G +V +W+ + W L
Sbjct: 359 IATLEAPPSEVWQMRFDPK----GAILAVAG-GGSASVNLWD----TSTWELVVTLSIPR 511
Query: 65 -------DETHTRT-VRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEV 116
D++ ++ V S AWSP GK LA S D T ++++ F + LEGH V
Sbjct: 512 VEGPKPSDKSGSKKFVLSVAWSPDGKRLACGSMDGTISVFDVQRAKF--LHHLEGHFMPV 685
Query: 117 KSVSWNA-SGTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILIS 175
+S+ ++ LL + S D +V +++ + + H V V P + +
Sbjct: 686 RSLVYSPYDPRLLFSASDDGNVHMYDAEGK---ALXGTMSXHASWVLCVDVSPNGAAIAT 856
Query: 176 CSYDNSIKVW 185
S D ++++W
Sbjct: 857 GSSDKTVRLW 886
>TC82717 weakly similar to GP|21539583|gb|AAM53344.1 putative coatomer
complex subunit {Arabidopsis thaliana}, partial (29%)
Length = 1118
Score = 57.4 bits (137), Expect = 5e-09
Identities = 42/150 (28%), Positives = 70/150 (46%), Gaps = 5/150 (3%)
Frame = +1
Query: 41 SGDKTVRIWEQDLSSGLWACKAVLDETHTRTVRSCAWSPSG-KLLATASFDATTAIW--E 97
S D+ +++W W+C E ++ V A++P A+AS D T IW +
Sbjct: 442 SDDQVLKLWNWKKG---WSCDETF-EGNSHYVMQVAFNPKDPSTFASASLDGTLKIWTID 609
Query: 98 NVGGDFECVATLEGHENEVKSVSWNASGT--LLATCSRDKSVWIWEMQPVNEFECVSVLQ 155
+ +F T EGH + V + S L + S D + +W+ N CV L+
Sbjct: 610 SSAPNF----TFEGHLKGMNCVDYFESNDKQYLLSGSDDYTAKVWDYDSKN---CVQTLE 768
Query: 156 GHTQDVKMVKWHPTEDILISCSYDNSIKVW 185
GH +V + HP I+I+ S D+++K+W
Sbjct: 769 GHKNNVTAICAHPEIPIIITASEDSTVKIW 858
Score = 53.5 bits (127), Expect = 7e-08
Identities = 39/134 (29%), Positives = 64/134 (47%), Gaps = 2/134 (1%)
Frame = +1
Query: 10 QRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETHT 69
+ EG++ V + +NP P FAS S D T++IW D S+ + E H
Sbjct: 496 ETFEGNSHYVMQVAFNPKD-----PSTFASASLDGTLKIWTIDSSAPNFTF-----EGHL 645
Query: 70 RTVRSCAWSPSG--KLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGTL 127
+ + + S + L + S D T +W+ + CV TLEGH+N V ++ + +
Sbjct: 646 KGMNCVDYFESNDKQYLLSGSDDYTAKVWDYDSKN--CVQTLEGHKNNVTAICAHPEIPI 819
Query: 128 LATCSRDKSVWIWE 141
+ T S D +V IW+
Sbjct: 820 IITASEDSTVKIWD 861
Score = 33.5 bits (75), Expect = 0.073
Identities = 26/91 (28%), Positives = 40/91 (43%)
Frame = +1
Query: 39 SCSGDKTVRIWEQDLSSGLWACKAVLDETHTRTVRSCAWSPSGKLLATASFDATTAIWEN 98
S S D T ++W+ D + C L E H V + P ++ TAS D+T IW+
Sbjct: 700 SGSDDYTAKVWDYDSKN----CVQTL-EGHKNNVTAICAHPEIPIIITASEDSTVKIWDA 864
Query: 99 VGGDFECVATLEGHENEVKSVSWNASGTLLA 129
V + TL+ V S+ + + LA
Sbjct: 865 V--TYRLQNTLDFGLERVWSIGYKKGSSQLA 951
>TC79342 similar to GP|10177096|dbj|BAB10430. Notchless protein homolog
{Arabidopsis thaliana}, partial (47%)
Length = 873
Score = 57.4 bits (137), Expect = 5e-09
Identities = 40/136 (29%), Positives = 59/136 (42%), Gaps = 5/136 (3%)
Frame = +3
Query: 12 LEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETHTRT 71
+ GH + V S+ ++P G L AS SGD TVR W+ + ++ C H
Sbjct: 483 ISGHGEAVLSVAFSPD----GRQL--ASGSGDTTVRFWDLGTQTPMYTCTG-----HKNW 629
Query: 72 VRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSW-----NASGT 126
V AWSP GK L + S W+ G + L GH+ + +SW NA
Sbjct: 630 VLCIAWSPDGKYLVSGSMSGELICWDPQTGK-QLGNALTGHKKWITGISWEPVHLNAPCR 806
Query: 127 LLATCSRDKSVWIWEM 142
+ S+D IW++
Sbjct: 807 RFVSASKDGDARIWDV 854
Score = 55.1 bits (131), Expect = 2e-08
Identities = 36/131 (27%), Positives = 57/131 (43%), Gaps = 5/131 (3%)
Frame = +3
Query: 60 CKAVLDETHTRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSV 119
C A + H V S A+SP G+ LA+ S D T W+ G + T GH+N V +
Sbjct: 471 CSATISG-HGEAVLSVAFSPDGRQLASGSGDTTVRFWDL--GTQTPMYTCTGHKNWVLCI 641
Query: 120 SWNASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTE-----DILI 174
+W+ G L + S + W+ Q + + L GH + + + W P +
Sbjct: 642 AWSPDGKYLVSGSMSGELICWDPQTGKQLG--NALTGHKKWITGISWEPVHLNAPCRRFV 815
Query: 175 SCSYDNSIKVW 185
S S D ++W
Sbjct: 816 SASKDGDARIW 848
Score = 45.4 bits (106), Expect = 2e-05
Identities = 32/113 (28%), Positives = 48/113 (42%), Gaps = 1/113 (0%)
Frame = +3
Query: 105 CVATLEGHENEVKSVSWNASGTLLATCSRDKSVWIWEM-QPVNEFECVSVLQGHTQDVKM 163
C AT+ GH V SV+++ G LA+ S D +V W++ + C GH V
Sbjct: 471 CSATISGHGEAVLSVAFSPDGRQLASGSGDTTVRFWDLGTQTPMYTCT----GHKNWVLC 638
Query: 164 VKWHPTEDILISCSYDNSIKVWADEGDSDDWQCVQTLGQPNNGHTSTVWALSF 216
+ W P L+S S + W D Q + LG GH + +S+
Sbjct: 639 IAWSPDGKYLVSGSMSGELICW-------DPQTGKQLGNALTGHKKWITGISW 776
>TC79322 similar to GP|13605815|gb|AAK32893.1 At2g32700/F24L7.16
{Arabidopsis thaliana}, partial (59%)
Length = 1824
Score = 56.2 bits (134), Expect = 1e-08
Identities = 40/142 (28%), Positives = 62/142 (43%), Gaps = 9/142 (6%)
Frame = +2
Query: 75 CAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGTLLATCSRD 134
C +S GKLLA+A D +W + +T E H + V + + T LAT S D
Sbjct: 818 CHFSSDGKLLASAGHDKKVVLWNM--ETLKTQSTPEEHTVIITDVRFRPNSTQLATSSFD 991
Query: 135 KSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTE-DILISCSYDNSIKVWADEGDSDD 193
+V +W+ + + GHT V + +HP + D+ SC +N I+ W +
Sbjct: 992 TTVRLWD--AADPSVSLQAYSGHTSHVASLDFHPKKNDLFCSCDDNNEIRFW----NISQ 1153
Query: 194 WQC--------VQTLGQPNNGH 207
+ C Q QP +GH
Sbjct: 1154YSCTRVFKGGSTQVRFQPRSGH 1219
Score = 54.3 bits (129), Expect = 4e-08
Identities = 47/178 (26%), Positives = 80/178 (44%), Gaps = 1/178 (0%)
Frame = +2
Query: 9 VQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDETH 68
+Q GH V SLD++P +F SC + +R W + ++C V
Sbjct: 1034 LQAYSGHTSHVASLDFHPKKND-----LFCSCDDNNEIRFW----NISQYSCTRVFKGGS 1186
Query: 69 TRTVRSCAWSP-SGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGTL 127
T+ + P SG LLA AS + + +V D + + +L+GH EV V W+ +G
Sbjct: 1187 TQV----RFQPRSGHLLAAASGNVVSLF--DVETDRQ-MHSLQGHSGEVHCVCWDTNGDY 1345
Query: 128 LATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVW 185
LA+ S++ SV +W + +C+ L +HP+ L+ S+++W
Sbjct: 1346 LASVSQE-SVKVWSLA---SGDCIHELNSSGNMFHSCVFHPSYSNLLVIGGYQSLELW 1507
Score = 53.1 bits (126), Expect = 9e-08
Identities = 42/193 (21%), Positives = 81/193 (41%), Gaps = 1/193 (0%)
Frame = +2
Query: 36 VFASCSGDKTVRIWEQDLSSGLWACKAVLDETHTRTVRSCAWSPSGKLLATASFDATTAI 95
+ AS DK V +W + E HT + + P+ LAT+SFD T +
Sbjct: 842 LLASAGHDKKVVLWNMETLK-----TQSTPEEHTVIITDVRFRPNSTQLATSSFDTTVRL 1006
Query: 96 WENVGGDFECVATLEGHENEVKSVSWN-ASGTLLATCSRDKSVWIWEMQPVNEFECVSVL 154
W+ A GH + V S+ ++ L +C + + W ++++ C V
Sbjct: 1007 WDAADPSVSLQA-YSGHTSHVASLDFHPKKNDLFCSCDDNNEIRFWN---ISQYSCTRVF 1174
Query: 155 QGHTQDVKMVKWHPTEDILISCSYDNSIKVWADEGDSDDWQCVQTLGQPNNGHTSTVWAL 214
+G + V++ P L++ + N + ++ E D + +L GH+ V +
Sbjct: 1175 KGGS---TQVRFQPRSGHLLAAASGNVVSLFDVETDRQ----MHSL----QGHSGEVHCV 1321
Query: 215 SFNASGDKMVTCS 227
++ +GD + + S
Sbjct: 1322 CWDTNGDYLASVS 1360
Score = 50.1 bits (118), Expect = 7e-07
Identities = 34/131 (25%), Positives = 60/131 (44%), Gaps = 1/131 (0%)
Frame = +2
Query: 100 GGDFECVATLEGHENEVKSVSWNASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQ 159
G F V+++ +V +++ G LLA+ DK V +W M+ + + S + HT
Sbjct: 761 GFSFSEVSSIRKSNGKVVCCHFSSDGKLLASAGHDKKVVLWNMETL---KTQSTPEEHTV 931
Query: 160 DVKMVKWHPTEDILISCSYDNSIKVWADEGDSDDWQCVQTLGQPNNGHTSTVWALSFN-A 218
+ V++ P L + S+D ++++W S Q +GHTS V +L F+
Sbjct: 932 IITDVRFRPNSTQLATSSFDTTVRLWDAADPSVSLQAY-------SGHTSHVASLDFHPK 1090
Query: 219 SGDKMVTCSSN 229
D +C N
Sbjct: 1091KNDLFCSCDDN 1123
Score = 35.8 bits (81), Expect = 0.015
Identities = 33/135 (24%), Positives = 64/135 (46%)
Frame = +2
Query: 7 KEVQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDE 66
+++ L+GH+ V + W+ + AS S ++V++W L+SG C L+
Sbjct: 1274 RQMHSLQGHSGEVHCVCWDTNGDY------LASVS-QESVKVWS--LASG--DCIHELNS 1420
Query: 67 THTRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGT 126
+ SC + PS L + +W + +C+ T+ HE + +++ +
Sbjct: 1421 SGNM-FHSCVFHPSYSNLLVIGGYQSLELWHMA--ENKCM-TIPAHEGVISALAQSPVTG 1588
Query: 127 LLATCSRDKSVWIWE 141
++A+ S DKSV IW+
Sbjct: 1589 MVASASHDKSVKIWK 1633
>TC88475 similar to GP|20260442|gb|AAM13119.1 unknown protein {Arabidopsis
thaliana}, partial (96%)
Length = 1315
Score = 55.8 bits (133), Expect = 1e-08
Identities = 30/90 (33%), Positives = 50/90 (55%), Gaps = 2/90 (2%)
Frame = +3
Query: 111 GHENEVKSVSWNASGTLLATCSRDKSVWIWEMQPVNEFECVSV-LQGHTQDVKMVKWHPT 169
GH+ +V SV+WN GT LA+ S D++ IW + P + + L+GHT V + W P
Sbjct: 195 GHKKKVHSVAWNCIGTKLASGSVDQTARIWHIDPHAHGKVKDIELKGHTDSVDQLCWDPK 374
Query: 170 E-DILISCSYDNSIKVWADEGDSDDWQCVQ 198
D++ + S D ++++W D+ +C Q
Sbjct: 375 HPDLIATASGDKTVRLW----DARSGKCSQ 452
Score = 53.5 bits (127), Expect = 7e-08
Identities = 57/233 (24%), Positives = 100/233 (42%), Gaps = 3/233 (1%)
Frame = +3
Query: 5 ELKEVQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVL 64
++K+++ L+GH D V L W+P P + A+ SGDKTVR+W D SG + +A L
Sbjct: 309 KVKDIE-LKGHTDSVDQLCWDPKH-----PDLIATASGDKTVRLW--DARSGKCSQQAEL 464
Query: 65 DETHTRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNAS 124
+ + + P G +A + D I + F+ + + EV ++WN +
Sbjct: 465 SGENI----NITYKPDGTHVAVGNRDDELTILD--VRKFKPMHRRK-FNYEVNEIAWNMT 623
Query: 125 GTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKV 184
G + + + +V E+ + L HT + PT S D+ + +
Sbjct: 624 GEMFFLTTGNGTV---EVLSYPSLRPLDTLMAHTAGCYCIAIDPTGRHFAVGSADSLVSL 794
Query: 185 WADEGDSDDWQCVQTLGQPNNGHTSTVW---ALSFNASGDKMVTCSSNLLMAV 234
W + CV+T T W +SFN +GD + + S +L + +
Sbjct: 795 WV----ISEMLCVRTF-------TKLEWPVRTISFNHTGDLIASASEDLFIDI 920
Score = 45.1 bits (105), Expect = 2e-05
Identities = 27/78 (34%), Positives = 41/78 (51%), Gaps = 4/78 (5%)
Frame = +3
Query: 68 HTRTVRSCAWSPSGKLLATASFDATTAIWE---NVGGDFECVATLEGHENEVKSVSWNAS 124
H + V S AW+ G LA+ S D T IW + G + + L+GH + V + W+
Sbjct: 198 HKKKVHSVAWNCIGTKLASGSVDQTARIWHIDPHAHGKVKDI-ELKGHTDSVDQLCWDPK 374
Query: 125 -GTLLATCSRDKSVWIWE 141
L+AT S DK+V +W+
Sbjct: 375 HPDLIATASGDKTVRLWD 428
>AL383039 weakly similar to PIR|A45442|A454 transport versicle formation
protein SEC13 [validated] - yeast (Saccharomyces
cerevisiae), partial (16%)
Length = 348
Score = 55.5 bits (132), Expect = 2e-08
Identities = 31/74 (41%), Positives = 43/74 (57%), Gaps = 1/74 (1%)
Frame = +2
Query: 38 ASCSGDKTVRIWEQDLSSGLWACKAVLDETHTRTVRSCAWSPSGKLLATASFDATTAIW- 96
ASCS DKTV IW+++ + W K + E V S +WS SG +LA AS D +W
Sbjct: 29 ASCSQDKTVIIWKKNSPTSPWVKKFLKPEKFLDVVWSVSWSLSGTILAVASGDNKVTLWQ 208
Query: 97 ENVGGDFECVATLE 110
EN+ G++E V L+
Sbjct: 209 ENLKGEWEKVQELD 250
Score = 39.7 bits (91), Expect = 0.001
Identities = 27/74 (36%), Positives = 35/74 (46%), Gaps = 3/74 (4%)
Frame = +2
Query: 84 LATASFDATTAIWENVGGDFECVATL---EGHENEVKSVSWNASGTLLATCSRDKSVWIW 140
LA+ S D T IW+ V E + V SVSW+ SGT+LA S D V +W
Sbjct: 26 LASCSQDKTVIIWKKNSPTSPWVKKFLKPEKFLDVVWSVSWSLSGTILAVASGDNKVTLW 205
Query: 141 EMQPVNEFECVSVL 154
+ E+E V L
Sbjct: 206 QENLKGEWEKVQEL 247
Score = 30.8 bits (68), Expect = 0.47
Identities = 20/61 (32%), Positives = 32/61 (51%)
Frame = +2
Query: 6 LKEVQRLEGHNDRVWSLDWNPATGHAGTPLVFASCSGDKTVRIWEQDLSSGLWACKAVLD 65
+K+ + E D VWS+ W+ + GT + A SGD V +W+++L G W LD
Sbjct: 92 VKKFLKPEKFLDVVWSVSWSLS----GT--ILAVASGDNKVTLWQENL-KGEWEKVQELD 250
Query: 66 E 66
+
Sbjct: 251 D 253
Score = 30.0 bits (66), Expect = 0.80
Identities = 15/60 (25%), Positives = 29/60 (48%)
Frame = +2
Query: 170 EDILISCSYDNSIKVWADEGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVTCSSN 229
++ L SCS D ++ +W + W V+ +P VW++S++ SG + S +
Sbjct: 17 QNYLASCSQDKTVIIWKKNSPTSPW--VKKFLKPEK-FLDVVWSVSWSLSGTILAVASGD 187
>TC78016 similar to GP|21537191|gb|AAM61532.1 PRL1 protein {Arabidopsis
thaliana}, partial (81%)
Length = 1759
Score = 54.3 bits (129), Expect = 4e-08
Identities = 34/108 (31%), Positives = 52/108 (47%)
Frame = +1
Query: 68 HTRTVRSCAWSPSGKLLATASFDATTAIWENVGGDFECVATLEGHENEVKSVSWNASGTL 127
H VRS A PS AT S D T IW+ G + TL GH +V+ ++ + T
Sbjct: 583 HLGWVRSVAVDPSNTWFATGSADRTIKIWDLASGVLK--LTLTGHIEQVRGLAISHKHTY 756
Query: 128 LATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILIS 175
+ + DK V W+++ + + + GH V + HPT DIL++
Sbjct: 757 MFSAGDDKQVKCWDLE---QNKVIRSYHGHLSGVYCLAIHPTIDILLT 891
Score = 43.1 bits (100), Expect = 9e-05
Identities = 25/85 (29%), Positives = 41/85 (47%)
Frame = +3
Query: 106 VATLEGHENEVKSVSWNASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVK 165
+ L GHEN V SV + + T S D ++ +W+++ + +S L H + V+ +
Sbjct: 945 IHALSGHENTVCSVFTRPTDPQVVTGSHDSTIKMWDLR---YGKTMSTLTNHKKSVRAMA 1115
Query: 166 WHPTEDILISCSYDNSIKVWADEGD 190
HP E S S DN K +G+
Sbjct: 1116 QHPKEQAFASASADNIKKFTLPKGE 1190
Score = 39.7 bits (91), Expect = 0.001
Identities = 29/117 (24%), Positives = 55/117 (46%)
Frame = +1
Query: 109 LEGHENEVKSVSWNASGTLLATCSRDKSVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHP 168
+ GH V+SV+ + S T AT S D+++ IW++ L GH + V+ +
Sbjct: 574 ISGHLGWVRSVAVDPSNTWFATGSADRTIKIWDLA---SGVLKLTLTGHIEQVRGLAISH 744
Query: 169 TEDILISCSYDNSIKVWADEGDSDDWQCVQTLGQPNNGHTSTVWALSFNASGDKMVT 225
+ S D +K W D + + +++ +GH S V+ L+ + + D ++T
Sbjct: 745 KHTYMFSAGDDKQVKCW----DLEQNKVIRSY----HGHLSGVYCLAIHPTIDILLT 891
Score = 30.8 bits (68), Expect = 0.47
Identities = 22/94 (23%), Positives = 46/94 (48%), Gaps = 6/94 (6%)
Frame = +1
Query: 104 ECVATLEGHENEVKSVSWNASGTLLATCSRDK-SVWIWEMQPVNEFE-CVSVLQGHTQDV 161
E T+ ++ S+ W + +L D S+W W+ + + F+ +++Q + D
Sbjct: 1186 ENFVTICSLNRKLLSMQWLSMRRVLWLPGGDNGSMWFWDWKSGHNFQQSQTIVQPGSLDS 1365
Query: 162 KMVKWHPTEDI----LISCSYDNSIKVWADEGDS 191
+ + T D+ LISC D +IK+W ++ ++
Sbjct: 1366 EAGIYALTYDVTGTRLISCEADKTIKMWKEDDNA 1467
Score = 30.4 bits (67), Expect(2) = 5e-04
Identities = 14/50 (28%), Positives = 28/50 (56%)
Frame = +3
Query: 136 SVWIWEMQPVNEFECVSVLQGHTQDVKMVKWHPTEDILISCSYDNSIKVW 185
S +W+++ + + L GH V V PT+ +++ S+D++IK+W
Sbjct: 909 SAGVWDIRSKMQ---IHALSGHENTVCSVFTRPTDPQVVTGSHDSTIKMW 1049
Score = 29.3 bits (64), Expect(2) = 5e-04
Identities = 27/91 (29%), Positives = 38/91 (41%), Gaps = 12/91 (13%)
Frame = +1
Query: 19 VWSLDWNP---ATGHAG---------TPLVFASCSGDKTVRIWEQDLSSGLWACKAVLDE 66
VW W +GH G + FA+ S D+T++IW DL+SG+ K L
Sbjct: 541 VWHAPWKNYRVISGHLGWVRSVAVDPSNTWFATGSADRTIKIW--DLASGV--LKLTL-T 705
Query: 67 THTRTVRSCAWSPSGKLLATASFDATTAIWE 97
H VR A S + +A D W+
Sbjct: 706 GHIEQVRGLAISHKHTYMFSAGDDKQVKCWD 798
Score = 26.9 bits (58), Expect = 6.8
Identities = 10/21 (47%), Positives = 16/21 (75%)
Frame = +1
Query: 32 GTPLVFASCSGDKTVRIWEQD 52
GT L+ SC DKT+++W++D
Sbjct: 1402 GTRLI--SCEADKTIKMWKED 1458
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.129 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,661,405
Number of Sequences: 36976
Number of extensions: 162467
Number of successful extensions: 1124
Number of sequences better than 10.0: 140
Number of HSP's better than 10.0 without gapping: 763
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 972
length of query: 235
length of database: 9,014,727
effective HSP length: 93
effective length of query: 142
effective length of database: 5,575,959
effective search space: 791786178
effective search space used: 791786178
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0010.22


