

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
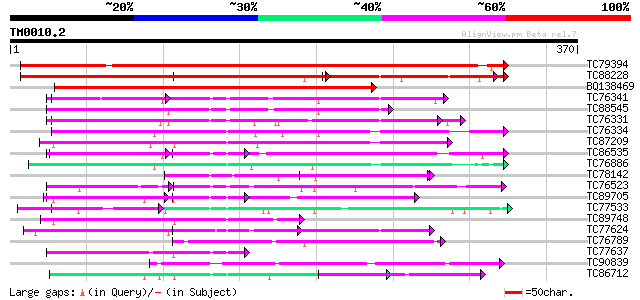
Query= TM0010.2
(370 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC79394 weakly similar to PIR|T49019|T49019 probable RNA binding... 504 e-143
TC88228 similar to GP|13605916|gb|AAK32943.1 AT4g00830/A_TM018A1... 257 1e-96
BQ138469 weakly similar to PIR|T49019|T49 probable RNA binding p... 305 2e-83
TC76341 similar to GP|7528270|gb|AAF63202.1| poly(A)-binding pro... 90 1e-18
TC88545 similar to GP|13560783|gb|AAK30205.1 poly(A)-binding pro... 87 1e-17
TC76331 similar to GP|7673355|gb|AAF66823.1| poly(A)-binding pro... 85 4e-17
TC76334 similar to GP|9663767|emb|CAC01237.1 RNA Binding Protein... 85 5e-17
TC87209 similar to GP|9663767|emb|CAC01237.1 RNA Binding Protein... 82 3e-16
TC86535 similar to GP|9758076|dbj|BAB08520.1 contains similarity... 74 9e-14
TC76886 similar to PIR|T01932|T01932 RNA binding protein homolog... 72 3e-13
TC78142 similar to GP|7578881|gb|AAF64167.1| plastid-specific ri... 71 8e-13
TC76523 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 70 1e-12
TC89705 similar to GP|21689785|gb|AAM67536.1 unknown protein {Ar... 61 6e-10
TC77533 similar to GP|6996560|emb|CAB75429.1 oligouridylate bind... 61 6e-10
TC89748 homologue to PIR|B84565|B84565 probable spliceosome asso... 60 1e-09
TC77624 similar to PIR|T12196|T12196 RNA-binding protein - fava ... 60 1e-09
TC76789 similar to GP|15294254|gb|AAK95304.1 AT4g24770/F22K18_30... 59 2e-09
TC77637 similar to GP|15292697|gb|AAK92717.1 putative RNA-bindin... 59 4e-09
TC90839 weakly similar to PIR|F86416|F86416 probable RNA-binding... 57 1e-08
TC86712 similar to GP|17528972|gb|AAL38696.1 putative RNA-bindin... 56 3e-08
>TC79394 weakly similar to PIR|T49019|T49019 probable RNA binding protein -
Arabidopsis thaliana, partial (74%)
Length = 1909
Score = 504 bits (1298), Expect = e-143
Identities = 254/324 (78%), Positives = 281/324 (86%), Gaps = 6/324 (1%)
Frame = +2
Query: 8 DEVEKKKHAELLALPPHSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERK 67
DE +KKKHAELLALPPH SEVYIGGI + SE+DLRVFCQSVGEV+EVR+MK KE+ K
Sbjct: 386 DEADKKKHAELLALPPHGSEVYIGGIPHETSEKDLRVFCQSVGEVAEVRVMKGKEA---K 556
Query: 68 GYAFVAFKTKELASQAIEELNNSEFKGKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVA 127
GYAFV FKTKELAS A++ELNNSEFKG+KIKCS SQ KHRLFIG++PK+WTVEDMKKVVA
Sbjct: 557 GYAFVTFKTKELASNALKELNNSEFKGRKIKCSPSQVKHRLFIGSVPKEWTVEDMKKVVA 736
Query: 128 DVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAE 187
VGPGVIS+ELLKD QSS RNRGFAFIEY+NHACAEYSRQKMSNSNFKL+NN VSWA+
Sbjct: 737 KVGPGVISVELLKDPQSSSRNRGFAFIEYHNHACAEYSRQKMSNSNFKLDNNDAIVSWAD 916
Query: 188 PRNSESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVH 247
PRNSESS+ SQVKAVYVKNLPENITQ+ LK+LFEHHGKITKV LPPAK+GQEKSR+GFVH
Sbjct: 917 PRNSESSSSSQVKAVYVKNLPENITQNRLKELFEHHGKITKVALPPAKAGQEKSRYGFVH 1096
Query: 248 FSDRSSAMKALKNTEKYEIDGKNLECSLAKPEADQRSSGTSNSQKPVVLPTYPHRLGYGM 307
F+DRSSAMKALKNTEKYEI+G+ LECSLAKP+ADQ+SSG SNS VLP YP LGYG
Sbjct: 1097FADRSSAMKALKNTEKYEINGQTLECSLAKPQADQKSSGASNSFNSAVLPAYPPPLGYG- 1273
Query: 308 VGGAYG------GIGAGYGAAGFA 325
YG +GAGYGAAGFA
Sbjct: 1274----YG*RWIWVAVGAGYGAAGFA 1333
>TC88228 similar to GP|13605916|gb|AAK32943.1 AT4g00830/A_TM018A10_14
{Arabidopsis thaliana}, partial (67%)
Length = 1712
Score = 257 bits (657), Expect(2) = 1e-96
Identities = 116/204 (56%), Positives = 164/204 (79%), Gaps = 2/204 (0%)
Frame = +1
Query: 8 DEVEKKKHAELLALPPHSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERK 67
DE E++KH ELL+ PPH SEV+IGG+ + S++D+R C+ +G++ E++++K +E+GE K
Sbjct: 409 DEEEREKHDELLSRPPHGSEVFIGGLPRDTSDDDVRELCEPMGDIVEIKLIKDRETGESK 588
Query: 68 GYAFVAFKTKELASQAIEELNNSEFKGKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVA 127
GYAFV +KTKE+A +AI++++N EFKGK ++C S+ KHRLFIGNIPK WT ++ +K V
Sbjct: 589 GYAFVGYKTKEVAQKAIDDIHNKEFKGKTLRCLLSETKHRLFIGNIPKTWTEDEFRKAVE 768
Query: 128 DVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAE 187
VGPGV SI+L+KD Q+ RNRGFAF+ YYN+ACA++SRQKMS+ FKL+ PTV+WA+
Sbjct: 769 GVGPGVESIDLIKDPQNQSRNRGFAFVLYYNNACADFSRQKMSSVGFKLDGITPTVTWAD 948
Query: 188 PRNS--ESSAVSQVKAVYVKNLPE 209
P+ S +S+A SQVKA+YVKN+PE
Sbjct: 949 PKTSPDQSAAASQVKALYVKNIPE 1020
Score = 114 bits (284), Expect(2) = 1e-96
Identities = 57/122 (46%), Positives = 82/122 (66%), Gaps = 1/122 (0%)
Frame = +2
Query: 205 KNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALKNTEKY 264
K N+T + LK+LF HG++TKVV+PP K+ ++ FGF+H+++RSSA+KA+K TEKY
Sbjct: 1007 KTYLRNVTTEQLKELFRRHGEVTKVVMPPGKASGKRD-FGFIHYAERSSALKAVKETEKY 1183
Query: 265 EIDGKNLECSLAKPEADQRSSGTSNSQKPVVLPTYPHRLGYG-MVGGAYGGIGAGYGAAG 323
EIDG+ LE +AKP+A+++ G + P + P + GYG G YG +GAGYG A
Sbjct: 1184 EIDGQALEVVIAKPQAEKKPDG-GYAYNPGLHPNHLPHPGYGNFSGNLYGSVGAGYGVAA 1360
Query: 324 FA 325
A
Sbjct: 1361 AA 1366
Score = 57.8 bits (138), Expect = 7e-09
Identities = 46/220 (20%), Positives = 104/220 (46%), Gaps = 8/220 (3%)
Frame = +1
Query: 108 LFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQ 167
+FIG +P+ + +D++++ +G ++ I+L+KD + +G ++G+AF+ Y A+ +
Sbjct: 469 VFIGGLPRDTSDDDVRELCEPMGD-IVEIKLIKD-RETGESKGYAFVGYKTKEVAQKAID 642
Query: 168 KMSNSNFKLENNAPTVSWAEPRNSESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGK-I 226
+ N FK + +S + R +++ N+P+ T+D +K E G +
Sbjct: 643 DIHNKEFKGKTLRCLLSETKHR------------LFIGNIPKTWTEDEFRKAVEGVGPGV 786
Query: 227 TKVVLPPAKSGQEKSR-FGFVHFSDRSSA--MKALKNTEKYEIDGKNLECSLAKPEADQR 283
+ L Q ++R F FV + + + A + ++ +++DG + A P+
Sbjct: 787 ESIDLIKDPQNQSRNRGFAFVLYYNNACADFSRQKMSSVGFKLDGITPTVTWADPKTSPD 966
Query: 284 SSGTSNSQKPVVLPTYPHRLGY----GMVGGAYGGIGAGY 319
S ++ K + + P + Y G + ++G +G+
Sbjct: 967 QSAAASQVKALYVKNIPEKCYY*TTKGAISSSWGSNESGH 1086
>BQ138469 weakly similar to PIR|T49019|T49 probable RNA binding protein -
Arabidopsis thaliana, partial (42%)
Length = 633
Score = 305 bits (781), Expect = 2e-83
Identities = 141/210 (67%), Positives = 180/210 (85%)
Frame = +2
Query: 30 IGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAIEELNN 89
+GGI + EDL+ FC+ +GEV +VRIMK K++ E KG+AFV ++ ELAS+AIEELNN
Sbjct: 2 VGGIPLDAKSEDLKEFCERIGEVVQVRIMKGKDASENKGFAFVTYRNVELASKAIEELNN 181
Query: 90 SEFKGKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNR 149
+EFKG+KIKCS+SQAK+RLFIGNIP+ W +D+KKVV D+GPGV ++EL+KD+++ NR
Sbjct: 182 TEFKGRKIKCSTSQAKNRLFIGNIPRSWGEKDLKKVVTDIGPGVTAVELVKDMKNISNNR 361
Query: 150 GFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAEPRNSESSAVSQVKAVYVKNLPE 209
GFAFI+Y+N+ CAEY RQKM + +FKL +N+PTVSWA+P+NS+SSA SQVKAVYVKNLP+
Sbjct: 362 GFAFIDYHNNQCAEYGRQKMMSPSFKLGDNSPTVSWADPKNSDSSASSQVKAVYVKNLPK 541
Query: 210 NITQDSLKKLFEHHGKITKVVLPPAKSGQE 239
N+TQ+ LKKLFEHHGKITKVVLPP KSGQE
Sbjct: 542 NVTQEQLKKLFEHHGKITKVVLPPPKSGQE 631
>TC76341 similar to GP|7528270|gb|AAF63202.1| poly(A)-binding protein
{Cucumis sativus}, partial (90%)
Length = 2501
Score = 90.1 bits (222), Expect = 1e-18
Identities = 71/274 (25%), Positives = 127/274 (45%), Gaps = 12/274 (4%)
Frame = +3
Query: 25 SSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAI 84
++ +Y+G + NV++ L VG+V VR+ + + GY +V F + A++A+
Sbjct: 291 TTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARAL 470
Query: 85 EELNNSEFKGKKIK--------CSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISI 136
+ LN + K I+ S +FI N+ K + + + G ++S
Sbjct: 471 DVLNFTPMNNKSIRVMYSHRDPSSRKSGTANIFIKNLDKTIDHKALHDTFSSFGQ-IMSC 647
Query: 137 ELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAEPRNSESSAV 196
++ D SG+++G+ F+++ A+ + K+ + + + V + + +
Sbjct: 648 KIATD--GSGQSKGYGFVQFEAEDSAQNAIDKL--NGMLINDKQVFVGHFLRKQDRDNVL 815
Query: 197 SQVK--AVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSA 254
S+ K VYVKNL E+ T+D LK F +G IT VL G+ K FGFV+F + A
Sbjct: 816 SKTKFNNVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDADGRSKC-FGFVNFENAEDA 992
Query: 255 MKALKNTEKYEIDGKNLECSLA--KPEADQRSSG 286
KA++ ++D K A K E +Q G
Sbjct: 993 AKAVEALNGKKVDDKEWYVGKAQKKSEREQELKG 1094
Score = 42.4 bits (98), Expect = 3e-04
Identities = 22/78 (28%), Positives = 43/78 (54%)
Frame = +3
Query: 28 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAIEEL 87
+Y+ + +++++E L+ G ++ +IM+ +G +G FVAF T E AS+A+ E+
Sbjct: 1149 LYLKNLDDSITDEKLKEMFSEFGTITSYKIMR-DPNGVSRGSGFVAFSTPEEASRALGEM 1325
Query: 88 NNSEFKGKKIKCSSSQAK 105
N K + + +Q K
Sbjct: 1326 NGKMIVSKPLYVAVAQRK 1379
>TC88545 similar to GP|13560783|gb|AAK30205.1 poly(A)-binding protein
{Daucus carota}, partial (35%)
Length = 866
Score = 87.0 bits (214), Expect = 1e-17
Identities = 65/237 (27%), Positives = 117/237 (48%), Gaps = 11/237 (4%)
Frame = +2
Query: 25 SSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAI 84
++ +Y+G + ++V++ L +G+V VRI + S + GY +V F A++A+
Sbjct: 176 TTSLYVGDLDHDVTDSQLYDLFNQIGQVVSVRICRDLASQQSLGYGYVNFSNPHDAAKAM 355
Query: 85 EELNNSEFKGKKIKCSSS--------QAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISI 136
+ LN + K I+ S +FI N+ + + + + G ++S
Sbjct: 356 DVLNFTPLNNKPIRIMYSHRDPSVRKSGAANIFIKNLDRAIDHKALYDTFSIFG-NILSC 532
Query: 137 ELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPT-VSWAEPRNSESSA 195
++ D +SG ++G+ F+++ N A+ + K+ N L N+ P V + + +A
Sbjct: 533 KIAMD--ASGLSKGYGFVQFENEESAQSAIDKL---NGMLLNDKPVYVGHFQRKQDRDNA 697
Query: 196 VSQVK--AVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSD 250
+S K VYVKNL E++T D LK F +G IT V+ G+ K FGFV+F +
Sbjct: 698 LSNAKFNNVYVKNLSESVTDDDLKNTFGEYGTITSAVVMRDVDGKSKC-FGFVNFEN 865
>TC76331 similar to GP|7673355|gb|AAF66823.1| poly(A)-binding protein
{Nicotiana tabacum}, partial (97%)
Length = 2581
Score = 85.1 bits (209), Expect = 4e-17
Identities = 65/268 (24%), Positives = 127/268 (47%), Gaps = 10/268 (3%)
Frame = +3
Query: 25 SSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAI 84
++ +Y+G + NV++ L +G+V VR+ + + GY +V + + A++A+
Sbjct: 441 TTSLYVGDLDMNVTDSQLYDLFNQLGQVVSVRVCRDLTTRRSLGYGYVNYSNPQDAARAL 620
Query: 85 EELNNSEFKGKKIKCSSS--------QAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISI 136
+ LN + + I+ S + +FI N+ K + + + G ++S
Sbjct: 621 DVLNFTPLNNRPIRIMYSHRDPSIRKSGQGNIFIKNLDKAIDHKALHDTFSSFG-NILSC 797
Query: 137 ELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNS--NFKLENNAPTVSWAEPRNSESS 194
++ D SG+++G+ F+++ A+ + +K++ N K P + E ++
Sbjct: 798 KVAVD--GSGQSKGYGFVQFDTEEAAQKAIEKLNGMLLNDKQVYVGPFLRKQERESTGDR 971
Query: 195 AVSQVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSA 254
A + V+VKNL E+ T D LKK F G IT V+ G+ K FGFV+F A
Sbjct: 972 A--KFNNVFVKNLSESTTDDELKKTFGEFGTITSAVVMRDGDGKSKC-FGFVNFESTDDA 1142
Query: 255 MKALKNTEKYEIDGKNLECSLAKPEADQ 282
+A++ +ID K A+ ++++
Sbjct: 1143ARAVEALNGKKIDDKEWYVGKAQKKSER 1226
Score = 80.1 bits (196), Expect = 1e-15
Identities = 72/296 (24%), Positives = 133/296 (44%), Gaps = 26/296 (8%)
Frame = +3
Query: 28 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAIEEL 87
++I + + + L S G + ++ SG+ KGY FV F T+E A +AIE+L
Sbjct: 714 IFIKNLDKAIDHKALHDTFSSFGNILSCKVA-VDGSGQSKGYGFVQFDTEEAAQKAIEKL 890
Query: 88 NNSEFKGKKI-----------KCSSSQAK-HRLFIGNIPKKWTVEDMKKVVADVGPGVIS 135
N K++ + + +AK + +F+ N+ + T +++KK + G + S
Sbjct: 891 NGMLLNDKQVYVGPFLRKQERESTGDRAKFNNVFVKNLSESTTDDELKKTFGEFGT-ITS 1067
Query: 136 IELLKDLQSSGRNRGFAFIEYYN-----HACAEYSRQKMSNSNF-------KLENNAPTV 183
+++D G+++ F F+ + + A + +K+ + + K E
Sbjct: 1068 AVVMRD--GDGKSKCFGFVNFESTDDAARAVEALNGKKIDDKEWYVGKAQKKSEREHELK 1241
Query: 184 SWAEPRNSESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRF 243
E E++ Q +YVKNL ++I + LK+LF +G IT + +G +
Sbjct: 1242 IKFEQSMKEAADKYQGANLYVKNLDDSIADEKLKELFSSYGTITSCKVMRDPNGVSRGS- 1418
Query: 244 GFVHFSDRSSAMKALKNTEKYEIDGKNLECSLAKPEADQRS--SGTSNSQKPVVLP 297
GFV FS A +AL + K L +LA+ + D+R+ +PV +P
Sbjct: 1419 GFVAFSTPEEASRALLEMNGKMVASKPLYVTLAQRKEDRRARLQAQFAQMRPVSMP 1586
Score = 34.3 bits (77), Expect = 0.079
Identities = 20/89 (22%), Positives = 41/89 (45%)
Frame = +3
Query: 201 AVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALKN 260
++YV +L N+T L LF G++ V + + + +G+V++S+ A +AL
Sbjct: 447 SLYVGDLDMNVTDSQLYDLFNQLGQVVSVRVCRDLTTRRSLGYGYVNYSNPQDAARALDV 626
Query: 261 TEKYEIDGKNLECSLAKPEADQRSSGTSN 289
++ + + + + R SG N
Sbjct: 627 LNFTPLNNRPIRIMYSHRDPSIRKSGQGN 713
>TC76334 similar to GP|9663767|emb|CAC01237.1 RNA Binding Protein 45
{Nicotiana plumbaginifolia}, partial (68%)
Length = 1579
Score = 84.7 bits (208), Expect = 5e-17
Identities = 70/326 (21%), Positives = 143/326 (43%), Gaps = 28/326 (8%)
Frame = +1
Query: 28 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAIEEL 87
++IG + + E L + GE++ V++++ K++ + +GY F+ F T+ A + ++
Sbjct: 235 LWIGDLQYWMDENYLYTCFGNTGELTSVKVIRNKQTSQSEGYGFIEFNTRATAERVLQTY 414
Query: 88 NNSEFK---------------GKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPG 132
N + G++ H +F+G++ T +++
Sbjct: 415 NGTIMPNGGQNYRLNWATFSAGERSSRQDDGPDHTIFVGDLAADVTDYLLQETFRARYNS 594
Query: 133 VISIELLKDLQSSGRNRGFAFIEYYNH-----ACAEYSRQKMSNSNFKL--ENNAPTVSW 185
V +++ D + +GR++G+ F+ + + A E S ++ N +
Sbjct: 595 VKGAKVVID-RLTGRSKGYGFVRFADEGEQMRAMTEMQGVLCSTRPMRIGPATNKNPAAT 771
Query: 186 AEPRNSESSAVSQVK------AVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQE 239
+P+ S + + Q + ++V NL N+T D L+++F +G++ V +P K
Sbjct: 772 TQPKASYNPSGGQSENDPNNTTIFVGNLDPNVTDDHLRQVFTQYGELVHVKIPSGK---- 939
Query: 240 KSRFGFVHFSDRSSAMKALKNTEKYEIDGKNLECSLAKPEADQRSSGTSNSQKPVVLPTY 299
R GFV FSDRS A +A++ + G+N+ S + +++++
Sbjct: 940 --RCGFVQFSDRSCAEEAIRVLNGTLLGGQNVRLSWGRTPSNKQTQQD------------ 1077
Query: 300 PHRLGYGMVGGAYGGIGAGYGAAGFA 325
P + GY GG YG GY G+A
Sbjct: 1078PTQGGYPAAGGYYGYAQGGYENYGYA 1155
>TC87209 similar to GP|9663767|emb|CAC01237.1 RNA Binding Protein 45
{Nicotiana plumbaginifolia}, partial (69%)
Length = 1504
Score = 82.4 bits (202), Expect = 3e-16
Identities = 68/302 (22%), Positives = 138/302 (45%), Gaps = 32/302 (10%)
Frame = +3
Query: 20 ALPPHSSE-----VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAF 74
A+PP SS ++IG + + E L GEV V++++ K++ + +GY F+ F
Sbjct: 225 AVPPPSSADEVKTLWIGDLQYWMDENYLYNCFSHTGEVGSVKVIRNKQTNQSEGYGFLEF 404
Query: 75 KTKELASQAIEELNNS-------EFKGKKIKCSSSQAKH------RLFIGNIPKKWTVED 121
++ A + ++ N + F+ SS + +H +F+G++ +
Sbjct: 405 ISRAGAERVLQTFNGTIMPNGGQNFRLNWATFSSGEKRHDDSPDYTIFVGDLAADVSDHH 584
Query: 122 MKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHA-------------CAEYSRQK 168
+ +V V +++ D +++GR +G+ F+ + + + C+ +
Sbjct: 585 LTEVFRTRYNSVKGAKVVID-RTTGRTKGYGFVRFADESEQMRAMTEMQGVLCSTRPMRI 761
Query: 169 MSNSNFKLENNAPTVSWAEPR-NSESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKIT 227
SN L S+ P+ +++ ++V NL N+T + LK++F +G++
Sbjct: 762 GPASNKNLGTQTSKASYQNPQGGAQNENDPNNTTIFVGNLDPNVTDEHLKQVFTQYGELV 941
Query: 228 KVVLPPAKSGQEKSRFGFVHFSDRSSAMKALKNTEKYEIDGKNLECSLAKPEADQRSSGT 287
V +P K R GFV F+DRSSA +AL+ + G+N+ S + A++++
Sbjct: 942 HVKIPSGK------RCGFVQFADRSSAEEALRVLNGTLLGGQNVRLSWGRSPANKQTQQD 1103
Query: 288 SN 289
N
Sbjct: 1104PN 1109
>TC86535 similar to GP|9758076|dbj|BAB08520.1 contains similarity to RNA
binding protein~gene_id:MNF13.1 {Arabidopsis thaliana},
partial (48%)
Length = 1555
Score = 73.9 bits (180), Expect = 9e-14
Identities = 55/228 (24%), Positives = 96/228 (41%), Gaps = 9/228 (3%)
Frame = +1
Query: 107 RLFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSR 166
++FIG + ++ T+ K G S+ ++KD + +G+ RGF FI Y + + +
Sbjct: 271 KIFIGGLARETTIAQFIKHFGKYGEITDSV-IMKD-RKTGQPRGFGFITYADPSVVD--- 435
Query: 167 QKMSNSNFKLENNAPTVSWAEPRNSESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKI 226
K+ + + + + PR + S + K ++V +P N+T+D + F +G++
Sbjct: 436 -KVIEDSHIINSKQVEIKRTIPRGAVGSKDFRTKKIFVGGIPSNVTEDEFRDFFTRYGEV 612
Query: 227 TKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALKNTEKYEIDGKNLECSLAKPEADQRSSG 286
+ S FGFV F + L K E G +E A+P+
Sbjct: 613 KDHQIMRDHSTNRSRGFGFVTFDTEEAVDDLLSMGNKIEFAGTQVEIKKAEPK------- 771
Query: 287 TSNSQKPVVLPTYPHRLGYGM---------VGGAYGGIGAGYGAAGFA 325
+N+ P R YG GG +GG+G GY +G A
Sbjct: 772 KANAPPPSSKRYNDSRSSYGSGGYGDAYDGFGGGFGGVGGGYSRSGSA 915
Score = 56.2 bits (134), Expect = 2e-08
Identities = 24/81 (29%), Positives = 46/81 (56%)
Frame = +1
Query: 25 SSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAI 84
+ ++++GGI +NV+E++ R F GEV + +IM+ + +G+ FV F T+E +
Sbjct: 529 TKKIFVGGIPSNVTEDEFRDFFTRYGEVKDHQIMRDHSTNRSRGFGFVTFDTEEAVDDLL 708
Query: 85 EELNNSEFKGKKIKCSSSQAK 105
N EF G +++ ++ K
Sbjct: 709 SMGNKIEFAGTQVEIKKAEPK 771
Score = 52.0 bits (123), Expect = 4e-07
Identities = 32/138 (23%), Positives = 67/138 (48%), Gaps = 8/138 (5%)
Frame = +1
Query: 27 EVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAIEE 86
+++IGG+A + GE+++ IMK +++G+ +G+ F+ + + + IE+
Sbjct: 271 KIFIGGLARETTIAQFIKHFGKYGEITDSVIMKDRKTGQPRGFGFITYADPSVVDKVIED 450
Query: 87 LNNSEFKGKKIK--------CSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIEL 138
+ K +IK S ++F+G IP T ++ + G V ++
Sbjct: 451 SHIINSKQVEIKRTIPRGAVGSKDFRTKKIFVGGIPSNVTEDEFRDFFTRYGE-VKDHQI 627
Query: 139 LKDLQSSGRNRGFAFIEY 156
++D S+ R+RGF F+ +
Sbjct: 628 MRD-HSTNRSRGFGFVTF 678
>TC76886 similar to PIR|T01932|T01932 RNA binding protein homolog - common
tobacco (fragment), partial (67%)
Length = 1896
Score = 72.0 bits (175), Expect = 3e-13
Identities = 70/358 (19%), Positives = 144/358 (39%), Gaps = 45/358 (12%)
Frame = +2
Query: 13 KKHAELLALPPHSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFV 72
++H + P +++G + + + E L GEV+ ++++ K++G+ +GY FV
Sbjct: 509 QQHQQKQLAPEEIRTIWLGDLHHWMDETFLHNCFAHTGEVASAKVIRNKQTGQSEGYGFV 688
Query: 73 AFKTKELASQAIEELNNSEFK------------------GKKIKCSSSQAKHRLFIGNIP 114
F T+ +A + ++ N + G + + S + + +F+G++
Sbjct: 689 EFYTRAMAEKVLQNFNGTMMPNTDQAFRLNWATFSAAGGGGERRSSEATSDLSVFVGDLA 868
Query: 115 KKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEY-----YNHACAEYSRQKM 169
T +++ A + +++ D ++GR++G+ F+ + A E +
Sbjct: 869 IDVTDAMLQETFASKFSSIKGAKVVID-SNTGRSKGYGFVRFGDESERTRAMTEMNGVYC 1045
Query: 170 SNSNFKLENNAPTVSWAEPRNSESSAV----------------------SQVKAVYVKNL 207
S+ ++ P ++ P+ S AV S ++V L
Sbjct: 1046SSRPMRVGVATPKKTYGNPQQYSSQAVVLAGGGGHSNGAMAQGSQSEGDSNNTTIFVGGL 1225
Query: 208 PENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALKNTEKYEID 267
+I+ + L++ F G + V +P K GFV +DR +A +A++ I
Sbjct: 1226DSDISDEDLRQPFLQFGDVISVKIPVGKG------CGFVQLADRKNAEEAIQGLNGTVIG 1387
Query: 268 GKNLECSLAKPEADQRSSGTSNSQKPVVLPTYPHRLGYGMVGGAYGGIGAGYGAAGFA 325
+ + S + ++ SN GYG G YG G GYG+ G+A
Sbjct: 1388KQTVRLSWGRSPGNKHWRNDSN--------------GYG--GHGYG--GHGYGSNGYA 1507
>TC78142 similar to GP|7578881|gb|AAF64167.1| plastid-specific ribosomal
protein 2 precursor {Spinacia oleracea}, partial (65%)
Length = 819
Score = 70.9 bits (172), Expect = 8e-13
Identities = 50/182 (27%), Positives = 84/182 (45%), Gaps = 7/182 (3%)
Frame = +2
Query: 102 SQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHAC 161
S A +L++GNIP+ + ++++K+V + G V E++ D S R+R FAF+
Sbjct: 242 SPALRKLYVGNIPRTVSNDELEKIVQEHG-AVEKAEVMYDKYSK-RSRRFAFVTMKTVED 415
Query: 162 AEYSRQKMSNSNF-------KLENNAPTVSWAEPRNSESSAVSQVKAVYVKNLPENITQD 214
A + +K++ + + T + ES+ V VYV NL +N+T D
Sbjct: 416 ANAAAEKLNGTEIGGREIKVNITEKPLTTEGLPVQAGESTFVDSPYKVYVGNLAKNVTSD 595
Query: 215 SLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALKNTEKYEIDGKNLECS 274
SLKK F G + A + S FGFV FS A+ + ++G+ + +
Sbjct: 596 SLKKFFSEKGNALSAKVSRAPGTSKSSGFGFVTFSSDEDVEAAISSFNNALLEGQKIRVN 775
Query: 275 LA 276
A
Sbjct: 776 KA 781
Score = 44.7 bits (104), Expect = 6e-05
Identities = 23/90 (25%), Positives = 44/90 (48%), Gaps = 2/90 (2%)
Frame = +2
Query: 190 NSESSAVSQ--VKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVH 247
+SE + V ++ +YV N+P ++ D L+K+ + HG + K + K + RF FV
Sbjct: 218 SSEQATVDSPALRKLYVGNIPRTVSNDELEKIVQEHGAVEKAEVMYDKYSKRSRRFAFVT 397
Query: 248 FSDRSSAMKALKNTEKYEIDGKNLECSLAK 277
A A + EI G+ ++ ++ +
Sbjct: 398 MKTVEDANAAAEKLNGTEIGGREIKVNITE 487
>TC76523 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (69%)
Length = 1615
Score = 70.1 bits (170), Expect = 1e-12
Identities = 67/233 (28%), Positives = 100/233 (42%), Gaps = 16/233 (6%)
Frame = +3
Query: 108 LFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQ 167
LF+GN+ D++K D G V+ + D GR +GF +E+ A AE ++
Sbjct: 555 LFVGNLSFSVQRSDIEKFFQDCGE-VVDVRFSSD--EEGRFKGFGHVEF---ASAEAAQS 716
Query: 168 KMSNSNFKLENNAPTVSWAEPR-------NSESSAVS----QVKAVYVKNLPENITQDSL 216
+ + +L A + A R NS SA S Q + V+V+ +N+ +D +
Sbjct: 717 ALEMNGQELLQRAVRLDLARERGAFTPNNNSNYSAQSGGRGQSQTVFVRGFDKNLGEDEI 896
Query: 217 K-KLFEHHG----KITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALKNTEKYEIDGKNL 271
+ KL EH G + T+V +P F ++ F D S KAL+ E E+DG L
Sbjct: 897 RAKLMEHFGGTCGEPTRVSIPKDFESGYSKGFAYMDFKDSDSFSKALELHES-ELDGYQL 1073
Query: 272 ECSLAKPEADQRSSGTSNSQKPVVLPTYPHRLGYGMVGGAYGGIGAGYGAAGF 324
AKP Q S G + + R G G G +GG G G F
Sbjct: 1074SVDEAKPRDSQGSGGRGGGGR-----SGGGRFGGGGRSGGFGGRSGGRGGGRF 1217
Score = 45.1 bits (105), Expect = 4e-05
Identities = 31/88 (35%), Positives = 49/88 (55%), Gaps = 5/88 (5%)
Frame = +3
Query: 25 SSEVYIGGIANNVSEEDLRV-----FCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKEL 79
S V++ G N+ E+++R F + GE + V I K ESG KG+A++ FK +
Sbjct: 843 SQTVFVRGFDKNLGEDEIRAKLMEHFGGTCGEPTRVSIPKDFESGYSKGFAYMDFKDSDS 1022
Query: 80 ASQAIEELNNSEFKGKKIKCSSSQAKHR 107
S+A+ EL+ SE G ++ S +AK R
Sbjct: 1023FSKAL-ELHESELDGYQL--SVDEAKPR 1097
>TC89705 similar to GP|21689785|gb|AAM67536.1 unknown protein {Arabidopsis
thaliana}, partial (46%)
Length = 812
Score = 61.2 bits (147), Expect = 6e-10
Identities = 38/148 (25%), Positives = 81/148 (54%), Gaps = 14/148 (9%)
Frame = +3
Query: 23 PHSSE------VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKT 76
PHS + ++IGG+A + + E+ + + GE+++ IMK + +G +G+ F+ +
Sbjct: 84 PHSGDGASPGKIFIGGLAKDTTLEEFVKYFERYGEITDSVIMKDRHTGRPRGFGFITYAD 263
Query: 77 KELASQAIEE---LNNSEFKGKK-IKCSSSQAK----HRLFIGNIPKKWTVEDMKKVVAD 128
+ Q I+E +N+ + + K+ I +SQ ++F+G IP + +++K +
Sbjct: 264 ASVVDQVIQENHIINDKQVEIKRTIPKGASQTNDFKTKKIFVGGIPATVSEDELKIFFSK 443
Query: 129 VGPGVISIELLKDLQSSGRNRGFAFIEY 156
G V+ E+++D ++ R+RGF F+ +
Sbjct: 444 HG-NVVEHEIIRD-HTTKRSRGFGFVVF 521
Score = 56.6 bits (135), Expect = 1e-08
Identities = 40/161 (24%), Positives = 74/161 (45%)
Frame = +3
Query: 107 RLFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSR 166
++FIG + K T+E+ K G S+ ++KD + +GR RGF FI Y + + +
Sbjct: 114 KIFIGGLAKDTTLEEFVKYFERYGEITDSV-IMKD-RHTGRPRGFGFITYADASVVDQVI 287
Query: 167 QKMSNSNFKLENNAPTVSWAEPRNSESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKI 226
Q+ N K T+ P+ + + + K ++V +P +++D LK F HG +
Sbjct: 288 QENHIINDKQVEIKRTI----PKGASQTNDFKTKKIFVGGIPATVSEDELKIFFSKHGNV 455
Query: 227 TKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALKNTEKYEID 267
+ + + + FGFV F + L + ++D
Sbjct: 456 VEHEIIRDHTTKRSRGFGFVVFDSDKAVDNLLADGNMIDMD 578
Score = 42.4 bits (98), Expect = 3e-04
Identities = 21/80 (26%), Positives = 42/80 (52%)
Frame = +3
Query: 25 SSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAI 84
+ ++++GGI VSE++L++F G V E I++ + +G+ FV F + + +
Sbjct: 372 TKKIFVGGIPATVSEDELKIFFSKHGNVVEHEIIRDHTTKRSRGFGFVVFDSDKAVDNLL 551
Query: 85 EELNNSEFKGKKIKCSSSQA 104
+ N + +I C +S A
Sbjct: 552 ADGNMIDMDDTQI-CGTSHA 608
>TC77533 similar to GP|6996560|emb|CAB75429.1 oligouridylate binding protein
{Nicotiana plumbaginifolia}, partial (79%)
Length = 1621
Score = 61.2 bits (147), Expect = 6e-10
Identities = 85/359 (23%), Positives = 135/359 (36%), Gaps = 58/359 (16%)
Frame = +1
Query: 28 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAIEEL 87
VY+G I VSE L+ S G + ++++ E+ Y FV + + A+ AI L
Sbjct: 226 VYVGNIHPQVSEPLLQELFSSAGALEGCKLIRK----EKSSYGFVDYFDRSSAAIAIVTL 393
Query: 88 NNSEFKGKKIKCS---------SSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIEL 138
N G+ IK + + +F+G++ + T + + +
Sbjct: 394 NGRNIFGQSIKVNWAYTRGQREDTSGHFHIFVGDLSPEVTDATLYACFSAYS-SCSDARV 570
Query: 139 LKDLQSSGRNRGFAFIEYYNHACAEY-----------SRQ----------------KMSN 171
+ D Q +GR+RGF F+ + N A+ SRQ + S
Sbjct: 571 MWD-QKTGRSRGFGFVSFRNQQEAQSAINDLTGKWLGSRQIRCNWATKGANMNGENQSSE 747
Query: 172 SNFKLENNAPTVSWAEPRNSESSAVS--QVKAVYVKNLPENITQDSLKKLFEHHGKITKV 229
S +E + T A+ S+ S Q VYV NL +T L HH V
Sbjct: 748 SKSVVELTSGTSEEAQEMTSDDSPEKNPQYTTVYVGNLAPEVTSVDL----HHHFHALGV 915
Query: 230 VLPPAKSGQEKSRFGFVHFSDRSSAMKALKNTEKYEIDGKNLECSLAKPEADQRSSGT-- 287
Q FGFV +S A A++ + GK ++CS ++ T
Sbjct: 916 GTIEDVRVQRDKGFGFVRYSTHGEAALAIQMGNTRFLFGKPIKCSWGSKPTPPGTASTPL 1095
Query: 288 ---SNSQKPVV------LPTYPHRLGYGMVGGAY---------GGIGAGYGAAGFAQVS 328
+++ PV L Y +L + A+ G +GAGYG AGF V+
Sbjct: 1096PPPASTHVPVPGFSPAGLALYERQLALSKMNEAHAVKRAAMGMGALGAGYG-AGFPNVA 1269
Score = 43.1 bits (100), Expect = 2e-04
Identities = 29/97 (29%), Positives = 46/97 (46%), Gaps = 2/97 (2%)
Frame = +1
Query: 6 TNDEVEKKKHAELLALPPHSSEVYIGGIANNVSEEDLR--VFCQSVGEVSEVRIMKAKES 63
T++E ++ + P + VY+G +A V+ DL VG + +VR+ + K
Sbjct: 778 TSEEAQEMTSDDSPEKNPQYTTVYVGNLAPEVTSVDLHHHFHALGVGTIEDVRVQRDK-- 951
Query: 64 GERKGYAFVAFKTKELASQAIEELNNSEFKGKKIKCS 100
G+ FV + T A+ AI+ N GK IKCS
Sbjct: 952 ----GFGFVRYSTHGEAALAIQMGNTRFLFGKPIKCS 1050
Score = 39.3 bits (90), Expect = 0.002
Identities = 25/80 (31%), Positives = 43/80 (53%)
Frame = +1
Query: 197 SQVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMK 256
S ++VYV N+ +++ L++LF G + L +EKS +GFV + DRSSA
Sbjct: 211 STCRSVYVGNIHPQVSEPLLQELFSSAGALEGCKL----IRKEKSSYGFVDYFDRSSAAI 378
Query: 257 ALKNTEKYEIDGKNLECSLA 276
A+ I G++++ + A
Sbjct: 379 AIVTLNGRNIFGQSIKVNWA 438
>TC89748 homologue to PIR|B84565|B84565 probable spliceosome associated
protein [imported] - Arabidopsis thaliana, partial (56%)
Length = 707
Score = 60.5 bits (145), Expect = 1e-09
Identities = 52/185 (28%), Positives = 91/185 (49%), Gaps = 13/185 (7%)
Frame = +2
Query: 21 LPPHSSE------VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAF 74
L HS+E Y+G + +SEE L G V V + K + + + +GY FV F
Sbjct: 131 LGQHSAERNQDATAYVGNLDPQISEELLWELFVQAGPVVNVYVPKDRVTNQHQGYGFVEF 310
Query: 75 KTKELASQAIEELNNSEFKGKKIKCS-SSQAKH------RLFIGNIPKKWTVEDMKKVVA 127
+++E A AI+ LN + GK I+ + +SQ K LFIGN+ + + +
Sbjct: 311 RSEEDADYAIKVLNMIKLYGKPIRVNKASQDKKSLDVGANLFIGNLDPDVDEKLLYDTFS 490
Query: 128 DVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAE 187
G V + ++++D + +RGF FI Y + ++ + + M+ L N TVS+A
Sbjct: 491 AFGVIVTNPKIMRD-PDTRNSRGFGFISYDSFEASDSAIEAMNGQ--YLCNRQITVSYAY 661
Query: 188 PRNSE 192
++++
Sbjct: 662 KKDTK 676
>TC77624 similar to PIR|T12196|T12196 RNA-binding protein - fava bean
(fragment), partial (90%)
Length = 1214
Score = 60.5 bits (145), Expect = 1e-09
Identities = 45/195 (23%), Positives = 91/195 (46%), Gaps = 13/195 (6%)
Frame = +1
Query: 10 VEKKKH--AELLALPPHSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERK 67
VE+K++ E +A +++Y G + +V L + G + ++ +++G+ +
Sbjct: 337 VEEKENIGGETVAEVDTRTKLYFGNLPYSVDSALLAGLIEEYGSAELIEVLYDRDTGKSR 516
Query: 68 GYAFVAFKTKELASQAIEELNNSEFKGKKIKCSSS-----------QAKHRLFIGNIPKK 116
G+AFV E + I+ L+ EF G+ ++ + S + +++LF+GN+
Sbjct: 517 GFAFVTMSCVEDCNAVIQNLDGKEFMGRTLRVNFSDKPKPKEPLYPETEYKLFVGNLAWT 696
Query: 117 WTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKL 176
T E + + + G V+ +L D +G++RG+ F+ Y +E N +L
Sbjct: 697 VTSESLTQAFQEHGT-VVGARVLFD-GETGKSRGYGFVSYATK--SEMDTALAIMDNVEL 864
Query: 177 ENNAPTVSWAEPRNS 191
E VS A+ + S
Sbjct: 865 EGRTLRVSLAQGKRS 909
Score = 57.8 bits (138), Expect = 7e-09
Identities = 46/174 (26%), Positives = 88/174 (50%), Gaps = 3/174 (1%)
Frame = +1
Query: 107 RLFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSR 166
+L+ GN+P + ++ + G + IE+L D + +G++RGFAF+ +C E
Sbjct: 394 KLYFGNLPYSVDSALLAGLIEEYGSAEL-IEVLYD-RDTGKSRGFAFVTM---SCVEDCN 558
Query: 167 QKMSNSNFK-LENNAPTVSWAE-PRNSESSAVSQVKAVYVKNLPENITQDSLKKLFEHHG 224
+ N + K V++++ P+ E ++V NL +T +SL + F+ HG
Sbjct: 559 AVIQNLDGKEFMGRTLRVNFSDKPKPKEPLYPETEYKLFVGNLAWTVTSESLTQAFQEHG 738
Query: 225 KITKV-VLPPAKSGQEKSRFGFVHFSDRSSAMKALKNTEKYEIDGKNLECSLAK 277
+ VL ++G+ + +GFV ++ +S AL + E++G+ L SLA+
Sbjct: 739 TVVGARVLFDGETGKSRG-YGFVSYATKSEMDTALAIMDNVELEGRTLRVSLAQ 897
>TC76789 similar to GP|15294254|gb|AAK95304.1 AT4g24770/F22K18_30
{Arabidopsis thaliana}, partial (62%)
Length = 1294
Score = 59.3 bits (142), Expect = 2e-09
Identities = 46/182 (25%), Positives = 83/182 (45%), Gaps = 4/182 (2%)
Frame = +3
Query: 107 RLFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSR 166
++F+GN+P + V+ K G + I + + + R+RGF F+ E +
Sbjct: 456 KIFVGNLP--FDVDSEKLAQLFEQSGTVEIAEVIYNRDTDRSRGFGFVTMSTSEEVERAV 629
Query: 167 QKMSNSNFKLENNAPTVSWAEPRNS----ESSAVSQVKAVYVKNLPENITQDSLKKLFEH 222
K S F+L+ TV+ A PR + + + YV NLP ++ SL++LF
Sbjct: 630 NKFSG--FELDGRLLTVNKAAPRGTPRLRQPRTFNSGLRAYVGNLPWDVDNSSLEQLFSE 803
Query: 223 HGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALKNTEKYEIDGKNLECSLAKPEADQ 282
HGK+ + + FGFV S+ + A+ + +G+ + ++A+ E +
Sbjct: 804 HGKVESAQVVYDRETGRSRGFGFVTMSNEAEMNDAIAALDGQSFNGRAIRVNVAE-ERPR 980
Query: 283 RS 284
RS
Sbjct: 981 RS 986
>TC77637 similar to GP|15292697|gb|AAK92717.1 putative RNA-binding protein
{Arabidopsis thaliana}, partial (59%)
Length = 1724
Score = 58.5 bits (140), Expect = 4e-09
Identities = 40/154 (25%), Positives = 72/154 (45%), Gaps = 22/154 (14%)
Frame = +2
Query: 25 SSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAI 84
S +++IGGI+ + +EE LR + + GEV E IMK + +G +G+ FV F +A +
Sbjct: 383 SGKLFIGGISWDTNEERLREYFSTYGEVKEAVIMKDRTTGRARGFGFVVFIDPAVADIVV 562
Query: 85 EELNNSEFKGKKIKCSSSQAK----------------------HRLFIGNIPKKWTVEDM 122
+E +N + G+ ++ + + ++F+G + T D
Sbjct: 563 QEKHNID--GRMVEAKKAVPRDDQNVLSRTSGSIHGSPGPGRTRKIFVGGLASTVTESDF 736
Query: 123 KKVVADVGPGVISIELLKDLQSSGRNRGFAFIEY 156
KK G + + ++ D ++ R RGF FI Y
Sbjct: 737 KKYFDQFGT-ITDVVVMYD-HNTQRPRGFGFITY 832
>TC90839 weakly similar to PIR|F86416|F86416 probable RNA-binding protein
MEI2 36123-32976 [imported] - Arabidopsis thaliana,
partial (21%)
Length = 1178
Score = 57.0 bits (136), Expect = 1e-08
Identities = 51/232 (21%), Positives = 98/232 (41%)
Frame = +2
Query: 92 FKGKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGF 151
FKGK Q+ LF+GNI ++K + G ++ L ++ ++RGF
Sbjct: 254 FKGKS--SFGEQSSRTLFVGNITSNAEDSELKALFEQYGD-------IRTLYTACKHRGF 406
Query: 152 AFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAEPRNSESSAVSQVKAVYVKNLPENI 211
I YY+ A+ + + + N L + + ++ P+ + + + + L +
Sbjct: 407 VMISYYDLRAAQNAMKALQNRT--LSSRKLDIRYSIPKGNPTEKDIGHGTLMISGLDSAV 580
Query: 212 TQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALKNTEKYEIDGKNL 271
+D LK++F +G+I ++ P E + ++ F D A +L++ + GK++
Sbjct: 581 LKDELKRIFGFYGEIKEIYEYP-----EMNHIKYIEFYDVRGAEASLRSLNGICLAGKHI 745
Query: 272 ECSLAKPEADQRSSGTSNSQKPVVLPTYPHRLGYGMVGGAYGGIGAGYGAAG 323
+ P R T SQK P H L + G+ +G A+G
Sbjct: 746 KLEPGHPRNAIRM--TQPSQKGQDEPDLGHNLNDILFLRQKAGLSSGVIASG 895
>TC86712 similar to GP|17528972|gb|AAL38696.1 putative RNA-binding protein
{Arabidopsis thaliana}, partial (51%)
Length = 2144
Score = 55.8 bits (133), Expect = 3e-08
Identities = 57/249 (22%), Positives = 99/249 (38%), Gaps = 26/249 (10%)
Frame = +3
Query: 27 EVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELASQAIEE 86
+++IGGI+ + +E+ LR + Q+ G+V E IMK + +G +G+ FV F +A + + E
Sbjct: 186 KLFIGGISWDTNEDRLRQYFQNFGDVVEAVIMKDRTTGRARGFGFVVFADPSVAERVVME 365
Query: 87 ---LNNSEFKGKK--------IKCSSSQAKH-----------RLFIGNIPKKWTVEDMKK 124
++ + KK + S+ + H ++F+G + T D K
Sbjct: 366 KHVIDGRTVEAKKAVPRDDQNVFTRSNSSSHGSPAPTPIRTKKIFVGGLASTVTESDFKN 545
Query: 125 VVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQK----MSNSNFKLENNA 180
G + + ++ D ++ R RGF FI Y + E K ++ +++
Sbjct: 546 YFDQFGT-ITDVVVMYD-HNTQRPRGFGFITYDSEEAVEKVLHKTFHELNGKMVEVKRAV 719
Query: 181 PTVSWAEPRNSESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEK 240
P P + S V + Q L HG L PA G
Sbjct: 720 PKDLSPSPSRGQLGGFSYGTMSRVGSFSNGFAQGYNPSLIGGHGLRLDGRLSPANVG--- 890
Query: 241 SRFGFVHFS 249
R G+ FS
Sbjct: 891 -RSGYSLFS 914
Score = 47.4 bits (111), Expect = 9e-06
Identities = 31/109 (28%), Positives = 52/109 (47%)
Frame = +3
Query: 202 VYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALKNT 261
+++ + + +D L++ F++ G + + V+ ++ FGFV F+D S A + +
Sbjct: 189 LFIGGISWDTNEDRLRQYFQNFGDVVEAVIMKDRTTGRARGFGFVVFADPSVAERVV--M 362
Query: 262 EKYEIDGKNLECSLAKPEADQRSSGTSNSQKPVVLPTYPHRLGYGMVGG 310
EK+ IDG+ +E A P DQ SNS P R VGG
Sbjct: 363 EKHVIDGRTVEAKKAVPRDDQNVFTRSNSSSHGSPAPTPIRTKKIFVGG 509
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.132 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,675,503
Number of Sequences: 36976
Number of extensions: 159761
Number of successful extensions: 1552
Number of sequences better than 10.0: 162
Number of HSP's better than 10.0 without gapping: 1375
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1503
length of query: 370
length of database: 9,014,727
effective HSP length: 98
effective length of query: 272
effective length of database: 5,391,079
effective search space: 1466373488
effective search space used: 1466373488
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0010.2


