

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0010.19
(295 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
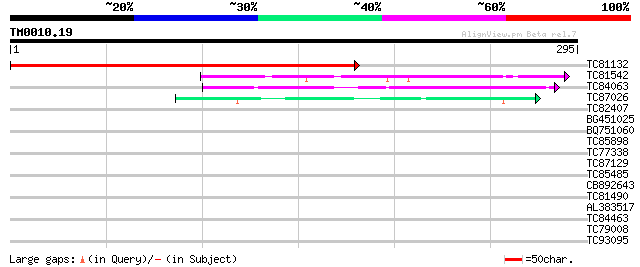
Score E
Sequences producing significant alignments: (bits) Value
TC81132 similar to GP|20466560|gb|AAM20597.1 unknown protein {Ar... 270 6e-73
TC81542 similar to GP|13877685|gb|AAK43920.1 Unknown protein {Ar... 103 1e-22
TC84063 similar to GP|7339697|dbj|BAA92902.1 hypothetical protei... 69 3e-12
TC87026 weakly similar to GP|21592576|gb|AAM64525.1 SOUL-like pr... 51 5e-07
TC82407 37 0.009
BG451025 similar to GP|20907905|gb| Transporter {Methanosarcina ... 30 0.85
BQ751060 similar to GP|14330324|emb aldo-keto reductase {Gallus ... 30 1.5
TC85898 similar to GP|12324950|gb|AAG52429.1 putative aminopepti... 29 2.5
TC77338 similar to GP|21536559|gb|AAM60891.1 adenine phosphoribo... 28 5.5
TC87129 weakly similar to PIR|F84503|F84503 hypothetical protein... 28 5.5
TC85485 similar to GP|22136900|gb|AAM91794.1 putative transketol... 28 5.5
CB892643 similar to PIR|B96582|B96 hypothetical protein F15I1.21... 28 5.5
TC81490 similar to GP|21592868|gb|AAM64818.1 unknown {Arabidopsi... 27 7.2
AL383517 similar to GP|21536749|gb| adenine phosphoribosyltransf... 27 7.2
TC84463 similar to PIR|S59634|S59634 endo-1 4-beta-xylanase (EC ... 27 7.2
TC79008 weakly similar to GP|18650596|gb|AAL75898.1 AT5g55220/MC... 27 7.2
TC93095 similar to GP|17473811|gb|AAL38336.1 unknown protein {Ar... 27 9.4
>TC81132 similar to GP|20466560|gb|AAM20597.1 unknown protein {Arabidopsis
thaliana}, partial (30%)
Length = 691
Score = 270 bits (689), Expect = 6e-73
Identities = 139/182 (76%), Positives = 157/182 (85%)
Frame = +1
Query: 1 MLPLCNASLSSQSSIRTSIPGRPNIAITNSASSSERISNRRRTMSAVEARSSLILALASQ 60
++PLCN SLS QS+ + P + I+ITNSASS+ R+S +RRT+SA EAR SLI ALASQ
Sbjct: 145 LIPLCNPSLSIQSAPISVKPNKTIISITNSASSNNRVSTQRRTISAFEARISLIFALASQ 324
Query: 61 ANSLTQRLVVDVATETARYLFPKRLESRTLEEALMAVPDLETVKFKVLSRRDNYEIREVE 120
+ SL+QRLV DVATETA+YLFPKR ESRTLEEALM VPDLETV FKVL+ RD YEIRE+E
Sbjct: 325 SFSLSQRLVADVATETAKYLFPKRFESRTLEEALMTVPDLETVNFKVLTIRDQYEIREIE 504
Query: 121 PYFVAEATMPGKSGFDFSGASQSFNVLAEYLFGKNTLKEKMEMTTPVYTSKNQSDGVKMD 180
PYFVAE TMPGKSGFDF G+SQSFNVLAEYLFGKNT KEKMEMTTPV+T+K QSDGVKMD
Sbjct: 505 PYFVAETTMPGKSGFDFRGSSQSFNVLAEYLFGKNTKKEKMEMTTPVFTTKKQSDGVKMD 684
Query: 181 MT 182
MT
Sbjct: 685 MT 690
>TC81542 similar to GP|13877685|gb|AAK43920.1 Unknown protein {Arabidopsis
thaliana}, partial (89%)
Length = 753
Score = 103 bits (256), Expect = 1e-22
Identities = 77/202 (38%), Positives = 103/202 (50%), Gaps = 10/202 (4%)
Frame = +3
Query: 100 LETVKFKVLSRRDNYEIREVEPYFVAEATMPGKSGFDFSGASQS-FNVLAEYLFG----K 154
+ET K++V +YEIR P AE T F G F VLA Y+ +
Sbjct: 3 VETPKYEVTKTTQDYEIRMYAPSVAAEVTYDPSQ---FKGNKDGGFMVLANYIGALGNPQ 173
Query: 155 NTLKEKMEMTTPVYTSKNQSDGVKMDMTTPVYTAKMEDQDK---WTMSFILPSKY--GAN 209
NT EK+ MT PV T + K+ MT PV T E+ ++ TM FILPS Y
Sbjct: 174 NTKPEKIAMTAPVIT---KGSAEKIAMTAPVVTKSSEEGERNKMVTMQFILPSSYEKAEE 344
Query: 210 LPLPKDSSVRIKEIPRKIVAVVSFSGFVNDEEVKQRELKLREALKSDGQFKIKKGTSVEV 269
P P D V I+E + VV FSG +DE VK++ KLR +L+ DG FK+ +
Sbjct: 345 APKPTDERVVIREEGERKYGVVKFSGVASDEVVKEKVEKLRLSLERDG-FKVI--GDFLL 515
Query: 270 AQYNPPFTLPFQRRNEIALEVE 291
+YNPP+TLP R NE+ + +E
Sbjct: 516 GRYNPPWTLPMFRTNEVMIPIE 581
>TC84063 similar to GP|7339697|dbj|BAA92902.1 hypothetical protein {Oryza
sativa (japonica cultivar-group)}, partial (42%)
Length = 599
Score = 68.6 bits (166), Expect = 3e-12
Identities = 58/189 (30%), Positives = 95/189 (49%), Gaps = 3/189 (1%)
Frame = +2
Query: 101 ETVKFKVLSRRDNYEIREVEPYFVAEATMPGKSGFDFSGASQS-FNVLAEYLFGKNTLKE 159
E+ ++ V+ ++EIR A P + F A+++ F+ L +++ G N
Sbjct: 80 ESPQYTVVHTESDFEIRLYRTSVWMSA--PAVNLISFEKATRNGFHRLFQFIQGANLNFS 253
Query: 160 KMEMTTPVYTSKNQSDGVKMDMTTPVYTAKMEDQDKWTMSFILPSKYGANLPLP-KDSSV 218
++ MTTPV T TT T +E Q + +SF LP+K+ N PLP + ++
Sbjct: 254 RIPMTTPVLT------------TTVPGTGPLESQGYY-VSFYLPTKFQENPPLPLPELNI 394
Query: 219 RIKEIPRKIVAVVSFSGFVNDEEVKQRELKLREAL-KSDGQFKIKKGTSVEVAQYNPPFT 277
+ VAV FSGF DE+V + KL +L + G+ ++K+ +AQYN PF
Sbjct: 395 KSYGFESHCVAVRGFSGFAKDEKVVKEAEKLDLSLSRWGGESEVKRKGGYSIAQYNGPFR 574
Query: 278 LPFQRRNEI 286
+ +RRNE+
Sbjct: 575 IA-KRRNEV 598
>TC87026 weakly similar to GP|21592576|gb|AAM64525.1 SOUL-like protein
{Arabidopsis thaliana}, partial (64%)
Length = 794
Score = 51.2 bits (121), Expect = 5e-07
Identities = 50/208 (24%), Positives = 81/208 (38%), Gaps = 18/208 (8%)
Frame = +1
Query: 87 SRTLEEALMAVPDLETVKFKVLSRRDNYEIR-----------EVEPYFVAEATMPGKSGF 135
S TL ++ +E + V+ + YEIR ++ + EAT G
Sbjct: 190 SGTLSYDIVPCKRIECPNYDVIEAGNGYEIRLYNSSVWISNSPIQDISLVEATRTG---- 357
Query: 136 DFSGASQSFNVLAEYLFGKNTLKEKMEMTTPVYTSKNQSDGVKMDMTTPVYTAKMEDQDK 195
F L +Y+ GKN ++K+EMT PV + SDG +
Sbjct: 358 --------FLRLFDYIQGKNNYQQKIEMTAPVLSEVLPSDGPFC-------------ESS 474
Query: 196 WTMSFILPSKYGANLPLPKDSSVRIKEIPRKIVAVVSFSGFVNDEEVKQRELKLREALKS 255
+ +SF +P AN P K + ++ AV F GFV D + + L++++
Sbjct: 475 FVVSFYVPKVNQANPPPAK--GLHVQRWKTVYAAVKQFGGFVKDTNIGEEAAALKDSIAG 648
Query: 256 -------DGQFKIKKGTSVEVAQYNPPF 276
+ + + VAQYN PF
Sbjct: 649 TKWSSAIEQSRRAGHASVYSVAQYNAPF 732
>TC82407
Length = 672
Score = 37.0 bits (84), Expect = 0.009
Identities = 32/128 (25%), Positives = 50/128 (39%), Gaps = 1/128 (0%)
Frame = +2
Query: 114 YEIREVEPYFVAEATMPGKSGFDFSGASQSFNVLAEYLFGKNTLKEKMEMTTPVYTSKNQ 173
Y++R P FV E MP ++ +++ +YL GKN K +E +PV
Sbjct: 257 YQLRLYNPRFVVE--MP------YTSRDKAYMAFDQYLTGKNDAKMSLETLSPVVMRHEP 412
Query: 174 SDGVK-MDMTTPVYTAKMEDQDKWTMSFILPSKYGANLPLPKDSSVRIKEIPRKIVAVVS 232
G K M M +++ P PLP D + + + V+V+
Sbjct: 413 DQGTKVMQMFVMPLGTRLQ-----------PEMLTQQPPLPDDPKLSLNIAGGEAVSVLR 559
Query: 233 FSGFVNDE 240
FSG E
Sbjct: 560 FSGQATKE 583
>BG451025 similar to GP|20907905|gb| Transporter {Methanosarcina mazei Goe1},
partial (3%)
Length = 422
Score = 30.4 bits (67), Expect = 0.85
Identities = 24/80 (30%), Positives = 42/80 (52%), Gaps = 2/80 (2%)
Frame = +3
Query: 218 VRIKEIPRKIVAVVSFSGFVNDEE--VKQRELKLREALKSDGQFKIKKGTSVEVAQYNPP 275
+ +KEIP+ V V++ V DE+ +K++EL LRE + + +F K + ++ Q
Sbjct: 96 IEVKEIPKTKVLVMAEMARVQDEQNKLKEKELNLRER-EFELEFLFKDISGMKARQ---- 260
Query: 276 FTLPFQRRNEIALEVERKDY 295
QR +E+ V R+ Y
Sbjct: 261 -----QRDHEMLCNVIREKY 305
>BQ751060 similar to GP|14330324|emb aldo-keto reductase {Gallus gallus},
partial (20%)
Length = 827
Score = 29.6 bits (65), Expect = 1.5
Identities = 22/82 (26%), Positives = 39/82 (46%), Gaps = 4/82 (4%)
Frame = +2
Query: 3 PLCNASLSSQSSIRTSIPGRPNIAITNSASSSERIS----NRRRTMSAVEARSSLILALA 58
P +++ +++ TS PG P+ + + A S +S T S+ EARSS +
Sbjct: 524 PSPTEAVTFSATLTTSRPGSPSRSSSRRARSVPLVSATTPRNTSTSSSPEARSSPPVNQI 703
Query: 59 SQANSLTQRLVVDVATETARYL 80
SL Q+ +VD+ E ++
Sbjct: 704 ENHPSLPQQEIVDLCREKGNHI 769
>TC85898 similar to GP|12324950|gb|AAG52429.1 putative aminopeptidase;
4537-10989 {Arabidopsis thaliana}, partial (90%)
Length = 3374
Score = 28.9 bits (63), Expect = 2.5
Identities = 25/85 (29%), Positives = 40/85 (46%), Gaps = 1/85 (1%)
Frame = -1
Query: 6 NASLSSQSSIRTSIPGRPNIA-ITNSASSSERISNRRRTMSAVEARSSLILALASQANSL 64
N L +SS + SI R I ++ +SSE ++NR+ S+V + S I AL
Sbjct: 2141 NGLLCWKSSTKLSINFRAKICPASHRLNSSESLANRKNRSSSVRSDSRRIGALYPLNKDG 1962
Query: 65 TQRLVVDVATETARYLFPKRLESRT 89
T + V T ++ +L R+ T
Sbjct: 1961 TGLSKISVNTNSSSFLVTLRIVVET 1887
>TC77338 similar to GP|21536559|gb|AAM60891.1 adenine
phosphoribosyltransferase 1 APRT {Arabidopsis
thaliana}, partial (74%)
Length = 1014
Score = 27.7 bits (60), Expect = 5.5
Identities = 24/82 (29%), Positives = 43/82 (52%), Gaps = 12/82 (14%)
Frame = +3
Query: 2 LPLCNASLSSQ-------SSIRTSIPG-----RPNIAITNSASSSERISNRRRTMSAVEA 49
L LC++S SS S++ ++P ++ ITNS SSS R ++ R T S + +
Sbjct: 24 LLLCSSSASSSNQLLLFGSAVTLTLPSCLRFPSLSLTITNSTSSSIRFNSIRSTSSQMAS 203
Query: 50 RSSLILALASQANSLTQRLVVD 71
+ S +A ++S+ R++ D
Sbjct: 204 KDSQDPRIARISSSI--RIIPD 263
>TC87129 weakly similar to PIR|F84503|F84503 hypothetical protein At2g12550
[imported] - Arabidopsis thaliana, partial (63%)
Length = 1894
Score = 27.7 bits (60), Expect = 5.5
Identities = 14/43 (32%), Positives = 26/43 (59%)
Frame = -1
Query: 55 LALASQANSLTQRLVVDVATETARYLFPKRLESRTLEEALMAV 97
LALA++ SL QRL++ + + T + P L++ + A+ A+
Sbjct: 895 LALAAEQTSLLQRLMLALISPTGQNEMPPPLQAALVSNAVQAL 767
>TC85485 similar to GP|22136900|gb|AAM91794.1 putative transketolase
{Arabidopsis thaliana}, partial (90%)
Length = 2728
Score = 27.7 bits (60), Expect = 5.5
Identities = 24/74 (32%), Positives = 37/74 (49%)
Frame = +2
Query: 7 ASLSSQSSIRTSIPGRPNIAITNSASSSERISNRRRTMSAVEARSSLILALASQANSLTQ 66
ASLSS SS+ S P S+SSS+ ++RRRT A+ A + + T+
Sbjct: 266 ASLSSTSSL--SFPA------LKSSSSSKPPTSRRRTTPAIRATA------VETLDKTTE 403
Query: 67 RLVVDVATETARYL 80
+V+ + T R+L
Sbjct: 404 ASLVEKSVNTIRFL 445
>CB892643 similar to PIR|B96582|B96 hypothetical protein F15I1.21 [imported]
- Arabidopsis thaliana, partial (35%)
Length = 758
Score = 27.7 bits (60), Expect = 5.5
Identities = 20/58 (34%), Positives = 26/58 (44%)
Frame = -3
Query: 28 TNSASSSERISNRRRTMSAVEARSSLILALASQANSLTQRLVVDVATETARYLFPKRL 85
TN SSE+I NR++ S ++ R S +L S TQR V T P L
Sbjct: 522 TNKIKSSEKIVNRKKLKSLIKHRRST*TSL-----SATQRETVPTTTVVTNKAPPNTL 364
>TC81490 similar to GP|21592868|gb|AAM64818.1 unknown {Arabidopsis
thaliana}, partial (27%)
Length = 754
Score = 27.3 bits (59), Expect = 7.2
Identities = 13/33 (39%), Positives = 21/33 (63%), Gaps = 1/33 (3%)
Frame = +2
Query: 203 PSKYGANLPLPKDSSVRIKEIPRKIV-AVVSFS 234
P ++ +NLPLP R K++PR I ++ +FS
Sbjct: 89 PLRFNSNLPLPPSFLRRRKQLPRSITKSLTAFS 187
>AL383517 similar to GP|21536749|gb| adenine phosphoribosyltransferase (EC
2.4.2.7)-like protein {Arabidopsis thaliana}, partial
(33%)
Length = 450
Score = 27.3 bits (59), Expect = 7.2
Identities = 22/75 (29%), Positives = 39/75 (51%), Gaps = 12/75 (16%)
Frame = +3
Query: 2 LPLCNASLSSQ-------SSIRTSIPG-----RPNIAITNSASSSERISNRRRTMSAVEA 49
L LC++S SS S++ ++P ++ ITNS SSS R ++ R T S + +
Sbjct: 12 LLLCSSSASSSNQLLLFGSAVTLTLPSCLRFPSLSLTITNSTSSSIRFNSIRSTSSQMAS 191
Query: 50 RSSLILALASQANSL 64
+ S +A ++S+
Sbjct: 192KDSQDPRIARISSSI 236
>TC84463 similar to PIR|S59634|S59634 endo-1 4-beta-xylanase (EC 3.2.1.8) F
precursor - Pseudomonas fluorescens, partial (4%)
Length = 1193
Score = 27.3 bits (59), Expect = 7.2
Identities = 24/76 (31%), Positives = 30/76 (38%)
Frame = -1
Query: 12 QSSIRTSIPGRPNIAITNSASSSERISNRRRTMSAVEARSSLILALASQANSLTQRLVVD 71
Q RTS P R + S+S RI+ + RSS A S + T R V
Sbjct: 572 QPRARTSCPARIPCRDPTTCSASRRITFSYTPAARTRLRSSAATAPRSSSCRNTSRSGVA 393
Query: 72 VATETARYLFPKRLES 87
+A L P R ES
Sbjct: 392 STISSAASLPPSRSES 345
>TC79008 weakly similar to GP|18650596|gb|AAL75898.1 AT5g55220/MCO15_17
{Arabidopsis thaliana}, partial (56%)
Length = 1365
Score = 27.3 bits (59), Expect = 7.2
Identities = 57/279 (20%), Positives = 104/279 (36%), Gaps = 18/279 (6%)
Frame = +1
Query: 9 LSSQSSIRTSIPGRPNIAITNSASSSERIS---NRRRTMSAVEARSSLILALASQANSLT 65
L +++ TS+P P+ ++ +S+SSS IS + T+ + S S + +
Sbjct: 43 LCTRTFTTTSLPFNPSFSLPSSSSSSSSISLHLHSNTTLKPFKFNLSSSFIQPSLYHHTS 222
Query: 66 QRLVVDVATETARYLFPKRLESRTLEEALMAVPDLET-------VKFKVLSRRDNYEIRE 118
V ++ ++ LFP E+ L L E+ V+ L D YE
Sbjct: 223 SPFTVSASSSSSVELFP---ENDRLPAQLKVTETKESNSRVTLHVEVPSLVCEDCYERVL 393
Query: 119 VEPYFVAEATMPGKSGFDFSGASQSFNVLAEYLFGKNTLKEKMEM--------TTPVYTS 170
VE F A +PG G ++L Y+ KN K +E T
Sbjct: 394 VE--FTKFAKVPGFR----PGKKIPESILVGYVGSKNVQKATIESILRRTLSHAVTAVTG 555
Query: 171 KNQSDGVKMDMTTPVYTAKMEDQDKWTMSFILPSKYGANLPLPKDSSVRIKEIPRKIVAV 230
+ D +++ K D + T S + KY ++ + + + + + V
Sbjct: 556 RALQDSIRI-------VTKFSDMED-TYSSLGYLKYDVSVDVAPEIKWISDDAYKNLKVV 711
Query: 231 VSFSGFVNDEEVKQRELKLREALKSDGQFKIKKGTSVEV 269
V ++ ++E + R KS G K+ ++V
Sbjct: 712 VEIDNEIDAHIASEKEFRRR--YKSAGVLKVVTDRGLQV 822
>TC93095 similar to GP|17473811|gb|AAL38336.1 unknown protein {Arabidopsis
thaliana}, partial (42%)
Length = 678
Score = 26.9 bits (58), Expect = 9.4
Identities = 11/20 (55%), Positives = 15/20 (75%)
Frame = +3
Query: 98 PDLETVKFKVLSRRDNYEIR 117
PDLE+ K+++L R NYE R
Sbjct: 618 PDLESPKYQILKRTANYEGR 677
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.129 0.347
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,538,778
Number of Sequences: 36976
Number of extensions: 68085
Number of successful extensions: 310
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 304
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 307
length of query: 295
length of database: 9,014,727
effective HSP length: 95
effective length of query: 200
effective length of database: 5,502,007
effective search space: 1100401400
effective search space used: 1100401400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0010.19


