

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0010.11
(1573 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
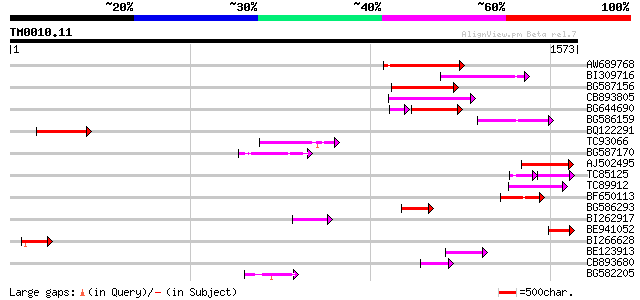
Score E
Sequences producing significant alignments: (bits) Value
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 275 1e-73
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 178 2e-44
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 166 5e-41
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 158 1e-38
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 130 3e-38
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 139 7e-33
BQ122291 133 5e-31
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 125 1e-28
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 120 4e-27
AJ502495 weakly similar to GP|18071369|g putative gag-pol polypr... 114 3e-25
TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-relate... 68 2e-23
TC89912 weakly similar to PIR|B84512|B84512 probable retroelemen... 99 1e-20
BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vu... 90 6e-18
BG586293 weakly similar to PIR|E84473|E84 probable retroelement ... 90 8e-18
BI262917 weakly similar to GP|19920130|g Putative retroelement {... 82 2e-15
BE941052 weakly similar to PIR|B85188|B85 retrotransposon like p... 79 2e-14
BI266628 78 3e-14
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 74 3e-13
CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotia... 73 1e-12
BG582205 54 5e-07
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 275 bits (703), Expect = 1e-73
Identities = 129/225 (57%), Positives = 171/225 (75%), Gaps = 1/225 (0%)
Frame = +1
Query: 1037 TRAKSGIVKPRLLPTLLLAQAEPVTTKHA-LKDPTWMAAMKSEYDALMSNNTWTLVTLPK 1095
TR K+G +KP++ L + P ++ L DP W+ AMK+EY AL+ N TW LV LP
Sbjct: 4 TRTKTGKLKPKVF----LTEPCPQDCENLPLSDPRWLQAMKTEYKALIDNKTWDLVPLPP 171
Query: 1096 GRHPIGCKWVFRIKENADGSVNRYKARLVAKGYHQVKGFDYAETFSPVVKPITIRLILTL 1155
+ IGCKWV+R+KEN DGSVN++KARLVAKG+ Q G DY ETFSPV+KP+TIRLILT+
Sbjct: 172 HKKAIGCKWVYRVKENPDGSVNKFKARLVAKGFSQTLGCDYTETFSPVIKPVTIRLILTI 351
Query: 1156 AVSKRWHIHQLDVNNAFLHGALQEEVYMQQPAGFQNSDKTLVCKLNKALYGLKQAPRAWF 1215
A++ +W I Q+D+NNAFL+G LQEEVYM QP GF+ ++K+LVCKLNK+LYGLKQAPRAW+
Sbjct: 352 AITYKWEIQQIDINNAFLNGFLQEEVYMSQPQGFEAANKSLVCKLNKSLYGLKQAPRAWY 531
Query: 1216 DRLKAALVKYGFKASCCDPSLFTFHNKSNVIYILVYVDDIIITGN 1260
+ L +A +++GF S CDPSL ++ IY+ +YVDDI+ITG+
Sbjct: 532 EXLTSAQIQFGFTKSRCDPSLLIYNQNGACIYLXIYVDDILITGS 666
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 178 bits (451), Expect = 2e-44
Identities = 99/246 (40%), Positives = 144/246 (58%)
Frame = +2
Query: 1195 TLVCKLNKALYGLKQAPRAWFDRLKAALVKYGFKASCCDPSLFTFHNKSNVIYILVYVDD 1254
T VC+L K++YGLKQA R W+ +L +L+ +G+ S D SLFT S+ +LVYVDD
Sbjct: 14 TKVCELQKSIYGLKQASRQWYSKLSESLISFGYLQSSSDFSLFTKFKDSSFTTLLVYVDD 193
Query: 1255 IIITGNFLPFIQEVVQKLNSEFALKQLGQLDYFLGVQVHHLQDRSLLLTQTKYIGDLLER 1314
I++ GN + IQ V L F +K LG L YFLG++V + +LL Q KY +LLE
Sbjct: 194 IVLAGNDISEIQHVKCFLIDRFKIKDLGSLRYFLGLEVAR-SKQGILLNQRKYTLELLED 370
Query: 1315 ADMAEAKGISTPMVSGARLSKHGADYFSDPTLYRSIVGALQYATLTRPEISFSVNKVCQF 1374
+ K TP +L + ++D T YR ++G L Y T TRP+ISF+V ++ QF
Sbjct: 371 SGNLAVKSTLTPYDISLKLHNSDSPLYNDETQYRRLIGKLIYLTTTRPDISFAVQQLSQF 550
Query: 1375 LSQPLEEHWKAVKRILRYLKGTIHHGLHIRPCSLHSPVSLVAYCDADWGSDPDDRRSTSG 1434
+S+P + H++A R+L+YLK GL S S + L ++ D+DW + P R+S +G
Sbjct: 551 VSKPQQVHYQAAIRVLQYLKTAPAKGLFY---SATSNLKLSSFADSDWATCPTTRKSVTG 721
Query: 1435 SCVFFG 1440
VF G
Sbjct: 722 YWVFLG 739
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 166 bits (421), Expect = 5e-41
Identities = 84/186 (45%), Positives = 120/186 (64%), Gaps = 1/186 (0%)
Frame = -1
Query: 1059 PVTTKHALKDPTWMAAMKSEYDALMSNNTWTLVTLPKGRHPIGCKWVFRIKENADGSVNR 1118
P + + A++D W ++ +E A++ N+TW LPKG+ + +W+F IK ADGS+ R
Sbjct: 558 PRSYEEAMEDKEWKESVGAEAGAMIKNDTWYESELPKGKKAVSSRWIFTIKYKADGSIER 379
Query: 1119 YKARLVAKGYHQVKGFDYAETFSPVVKPITIRLILTLAVSKRWHIHQLDVNNAFLHGALQ 1178
K RLVA+G+ G DY ETF+PV K TIR++L+LAV+ W + Q+DV NAFL G L+
Sbjct: 378 KKTRLVARGFTLTYGEDYIETFAPVAKLHTIRIVLSLAVNLGWGLWQMDVKNAFLQGELE 199
Query: 1179 EEVYMQQPAGFQNSDKT-LVCKLNKALYGLKQAPRAWFDRLKAALVKYGFKASCCDPSLF 1237
+EVYM P G ++ K V +L KA+YGLKQ+PRAW+++L L GF+ S D +LF
Sbjct: 198 DEVYMYPPPGLEHLVKRGNVLRLKKAIYGLKQSPRAWYNKLSTTLNGRGFRKSELDHTLF 19
Query: 1238 TFHNKS 1243
T S
Sbjct: 18 TLTTPS 1
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 158 bits (400), Expect = 1e-38
Identities = 90/244 (36%), Positives = 137/244 (55%), Gaps = 3/244 (1%)
Frame = +3
Query: 1052 LLLAQAEPVTTKHALKDPTWMAAMKSEYDALMSNNTWTLVTLPKGRHPIGCKWVFRIKEN 1111
+L ++P T + A+K W A+M +E +A NNTW L L G IG KW+F+ K N
Sbjct: 21 MLTMTSDPTTFEEAVKSEKWRASMNNEMEATERNNTWELTDLRSGAKTIGLKWIFKTKLN 200
Query: 1112 ADGSVNRYKARLVAKGYHQVKGFDYAETFSPVVKPITIRLILTLAVS-KRWHIHQLDVNN 1170
+G + +YKARLVAKGY Q G DY E F+PV + TIR+++ LA KR + +
Sbjct: 201 ENGEIEKYKARLVAKGYSQQYGVDYTEVFAPVARWDTIRMVIALAAQIKRDGVCIS*M*K 380
Query: 1171 AFLHGALQEEVYMQQPAGFQNSDKTLVC-KLNKALYGLKQAPRAWFDRLKAALVKYGFKA 1229
A H ++ + + + ++ ++ +ALYGLKQAPRAW+ R++A K GF+
Sbjct: 381 A--HSCMEN*MRKFLLINHRVM*RRVIS*RVKRALYGLKQAPRAWYSRIEAYFTKEGFEK 554
Query: 1230 SCCDPSLFT-FHNKSNVIYILVYVDDIIITGNFLPFIQEVVQKLNSEFALKQLGQLDYFL 1288
+ +LF ++ I +YVDD+I GN +E + + EF + LG++ YFL
Sbjct: 555 CPYEHTLFVKLSEGGKILIISLYVDDLIFIGNDENMFEEFKKSMKKEFNMSDLGKMHYFL 734
Query: 1289 GVQV 1292
GV+V
Sbjct: 735 GVEV 746
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 130 bits (328), Expect(2) = 3e-38
Identities = 64/141 (45%), Positives = 97/141 (68%), Gaps = 1/141 (0%)
Frame = -2
Query: 1116 VNRYKARLVAKGYHQVKGFDYAETFSPVVKPITIRLILTLAVSKRWHIHQLDVNNAFLHG 1175
+ R K++LV +GY+Q +G DY E FSPV + IR+++ A + ++Q+DV +AF++G
Sbjct: 424 ITRNKSKLVVQGYNQKEGIDYDEAFSPVARMEAIRILIAFAAFMGFKLYQMDVKSAFING 245
Query: 1176 ALQEEVYMQQPAGFQNSD-KTLVCKLNKALYGLKQAPRAWFDRLKAALVKYGFKASCCDP 1234
L+EEV+++QP GF++++ V +LNK LYGLKQAPRAW++RL L+K GFK D
Sbjct: 244 DLKEEVFVKQPPGFEDAEVPNHVFRLNKTLYGLKQAPRAWYERLSKFLLKNGFKRGKIDN 65
Query: 1235 SLFTFHNKSNVIYILVYVDDI 1255
+LF + ++ I VYVDDI
Sbjct: 64 TLFLLKRE*ELLIIQVYVDDI 2
Score = 48.1 bits (113), Expect(2) = 3e-38
Identities = 21/56 (37%), Positives = 31/56 (54%)
Frame = -3
Query: 1054 LAQAEPVTTKHALKDPTWMAAMKSEYDALMSNNTWTLVTLPKGRHPIGCKWVFRIK 1109
++ EP K AL+D W+ +M+ E + W LV P+G+ IG +WVFR K
Sbjct: 600 ISSIEPKNVKEALRDADWINSMQEELHQFERSKVWYLVPRPEGKTVIGTRWVFRNK 433
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 139 bits (351), Expect = 7e-33
Identities = 76/214 (35%), Positives = 122/214 (56%), Gaps = 1/214 (0%)
Frame = +1
Query: 1297 DRSLLLTQTKYIGDLLERADMAEAKGISTPMVSGARLSKHGADYFSDPTLYRSIVGALQY 1356
+ + + Q KY+ DLLER M ++ P+ +L K D T Y+ IVG L Y
Sbjct: 19 EEGIYICQRKYVTDLLERFGMEKSNLSRNPIAPRCKLIKDENGVKVDATKYKQIVGCLMY 198
Query: 1357 ATLTRPEISFSVNKVCQFLSQPLEEHWKAVKRILRYLKGTIHHGLHIRPCSLHSPVSLVA 1416
TRP++ + ++ + +F++ P E H AVKR+LRYL GTI+ G+ + + L A
Sbjct: 199 LAATRPDLMYVLSLISRFMNCPTELHMHAVKRVLRYLNGTINLGIMYK---RNGSEKLEA 369
Query: 1417 YCDADWGSDPDDRRSTSGSCVFFGPNLVTWTSKKQTLVARSSTEAEYRSLANTSAELLWI 1476
Y D+D+ D DDR+STSG V+W+SKKQ +V S+T+AE+ + A + + +W+
Sbjct: 370 YTDSDYAGDLDDRKSTSGYVFMLSSGAVSWSSKKQPVVTLSTTKAEFIAAAFCACQSVWM 549
Query: 1477 QSLLQELQVPFTVP-KVFCDNMGAVALTHNPVLH 1509
+ +L++L + ++CDN + L+ NPVLH
Sbjct: 550 RRVLEKLGYTQSGSITMYCDNNSTIKLSKNPVLH 651
>BQ122291
Length = 487
Score = 133 bits (335), Expect = 5e-31
Identities = 60/153 (39%), Positives = 99/153 (64%)
Frame = -2
Query: 74 DADRVNSVENPAFLVWEQQDSALFTWLLSSLSPSVLPTVVNCQHSWQIWEAVLDFFHAHS 133
DA R NPA+ WE +DS L TW+LS +SPS+L V +HSWQ+W+ + +
Sbjct: 486 DAAREAGTRNPAYTEWEDKDSLLCTWILSRISPSLLSRFVLLRHSWQVWDEIHSYCFTQM 307
Query: 134 LAQSTQLRSELRSITKGSKNTSDYLKRIKSIVNALISIGDPVTYREHLEAIFDGLPEDYS 193
+S QLRSELRSITKGS+ S+++ RI++I +L SIGDPV++R+ +E + + LPE++
Sbjct: 306 KTRSRQLRSELRSITKGSRTVSEFIARIRAISESLASIGDPVSHRDLIEVVLEALPEEFD 127
Query: 194 HLMTIVTSREFPCSISQAEAMVIAHEARLDRLR 226
++ V ++ S+ + E+ ++ E+R ++ +
Sbjct: 126 PIVASVNAKSEVVSLDELESQLLTQESRKEKFK 28
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 125 bits (315), Expect = 1e-28
Identities = 83/238 (34%), Positives = 116/238 (47%), Gaps = 15/238 (6%)
Frame = +1
Query: 692 LELIFADLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAMVELKF 751
L+ I +DLWGP+ + S G Y +T +D F R W+Y L+ K+ET F +++ +VE +
Sbjct: 97 LDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFKKWRILVETQT 276
Query: 752 GLKIKSVQTDGGTEF--KPLLPHFLKLGITHRLTCPHTHHQNGSVERKHRHIVETGLSLL 809
G +K + TD EF GI T P QNG ER R ++E +L
Sbjct: 277 GKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCML 456
Query: 810 ACANMPT--SYWDHAFLTATFLINRLPTPVLGNLSPYHKLFKVP--------ADYKILRV 859
+ A + W A TA L+NR P L FKVP DY LR+
Sbjct: 457 SNAGL*N*RDLWVEAASTACHLVNRSPHSALD--------FKVPEDIWSGNLVDYSNLRI 612
Query: 860 FGSACYPHLRPYNSSKLSFRSQECVFLGYSSSHKGYKCLASD---GRIYISKDVQFNE 914
FG Y + N KL+ R+ EC+FL Y+S KGY+ SD ++ +S+DV FNE
Sbjct: 613 FGCPAYALV---NDGKLAPRAGECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNE 777
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 120 bits (301), Expect = 4e-27
Identities = 77/209 (36%), Positives = 111/209 (52%), Gaps = 5/209 (2%)
Frame = -3
Query: 635 LWHSRLGHPHYEALQSALTTCKIPVPHKSKYTICHSCCVAKSHRLPSSASSTLYTAPLEL 694
LWH+RLGHPH AL L V ++K C +C + K + +ST+Y +L
Sbjct: 584 LWHARLGHPHGRALNLMLPG----VVFENKN--CEACILGKHCKNVFPRTSTVYENCFDL 423
Query: 695 IFADLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAMVELKFGLK 754
I+ DLW S+ S + YF+T +D S++TW+ L+ K F FQA V + K
Sbjct: 422 IYTDLWTAPSL-SRDNHKYFVTFIDEKSKYTWLTLIPSKDRVIDAFKNFQAYVTNHYHAK 246
Query: 755 IKSVQTDGGTE-----FKPLLPHFLKLGITHRLTCPHTHHQNGSVERKHRHIVETGLSLL 809
IK +++D G E FK L H GI H+ +CP+T QNG +RK++H++E SL+
Sbjct: 245 IKILRSDNGGEYTSYAFKSHLDHH---GILHQTSCPYTPQQNGVAKRKNKHLMEVARSLM 75
Query: 810 ACANMPTSYWDHAFLTATFLINRLPTPVL 838
AN+ TA +LIN +PT VL
Sbjct: 74 FQANVS---------TACYLINWIPTKVL 15
>AJ502495 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (9%)
Length = 542
Score = 114 bits (285), Expect = 3e-25
Identities = 52/146 (35%), Positives = 89/146 (60%), Gaps = 1/146 (0%)
Frame = +2
Query: 1420 ADWGSDPDDRRSTSGSCVFFGPNLVTWTSKKQTLVARSSTEAEYRSLANTSAELLWIQSL 1479
+DW D + R+STSG G ++W+SKKQ +VA S+ EAEY + + + + +W++ +
Sbjct: 2 SDWAGDTETRKSTSGYAFHLGTGAISWSSKKQPVVAFSTAEAEYIASTSCATQTVWLRRI 181
Query: 1480 LQELQVPFTVP-KVFCDNMGAVALTHNPVLHTRTKHMELDIFFVREKVQNQSLFVHHVPS 1538
L+ + P K++CDN A+AL+ NPV H R+KH+++ +RE + + + + + P+
Sbjct: 182 LEVMHHEQNTPTKIYCDNKSAIALSKNPVFHGRSKHIDIQFHKIRELIAEKEVVIEYCPT 361
Query: 1539 VDQIADIFTKALSPTRFEDLRSKLNV 1564
++IADIFTK L F L+ L +
Sbjct: 362 EEKIADIFTKPLKIESFYKLKKMLGM 439
>TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
(EC 3.4.23.-);, partial (7%)
Length = 705
Score = 67.8 bits (164), Expect(2) = 2e-23
Identities = 34/102 (33%), Positives = 56/102 (54%)
Frame = +3
Query: 1465 SLANTSAELLWIQSLLQELQVPFTVPKVFCDNMGAVALTHNPVLHTRTKHMELDIFFVRE 1524
SL E +W+Q L++EL V+CD+ A+ + NP H+RTKH+ + FVRE
Sbjct: 228 SLPQACKEAIWMQRLMEELGHKQEQITVYCDSQSALHIARNPAFHSRTKHIGIQYHFVRE 407
Query: 1525 KVQNQSLFVHHVPSVDQIADIFTKALSPTRFEDLRSKLNVRE 1566
V+ S+ + + + D +AD TK+++ +F RS + E
Sbjct: 408 VVEEGSVDMQKIHTNDNLADAMTKSINTDKFIWCRSSYGLLE 533
Score = 61.6 bits (148), Expect(2) = 2e-23
Identities = 34/78 (43%), Positives = 47/78 (59%)
Frame = +1
Query: 1386 VKRILRYLKGTIHHGLHIRPCSLHSPVSLVAYCDADWGSDPDDRRSTSGSCVFFGPNLVT 1445
VKRI+RY+KGT + C S +++ Y D+D+ D D R+ST+G V+
Sbjct: 1 VKRIMRYIKGTSG----VAVCFGGSELTVRGYVDSDFAGDHDKRKSTTGYVFTLAGGAVS 168
Query: 1446 WTSKKQTLVARSSTEAEY 1463
W SK QT+VA S+TEAEY
Sbjct: 169 WLSKLQTVVALSTTEAEY 222
>TC89912 weakly similar to PIR|B84512|B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(10%)
Length = 814
Score = 99.0 bits (245), Expect = 1e-20
Identities = 47/165 (28%), Positives = 93/165 (55%)
Frame = +1
Query: 1384 KAVKRILRYLKGTIHHGLHIRPCSLHSPVSLVAYCDADWGSDPDDRRSTSGSCVFFGPNL 1443
+A+K +L+YL ++ L + +L Y DAD+ + D R+S SG
Sbjct: 1 QALKWVLKYLNESLKSSLKYTKAAQEED-ALEGYVDADYAGNVDTRKSLSGFVFTLYGTT 177
Query: 1444 VTWTSKKQTLVARSSTEAEYRSLANTSAELLWIQSLLQELQVPFTVPKVFCDNMGAVALT 1503
++W + +Q++V S+T+AEY + + +W++ ++ EL + K+ CD+ A+ L
Sbjct: 178 ISWKANQQSVVTLSTTQAEYIAFVEGVKDAIWLKGMIGELGITQEYVKIHCDSQSAIHLA 357
Query: 1504 HNPVLHTRTKHMELDIFFVREKVQNQSLFVHHVPSVDQIADIFTK 1548
++ V H RTKH+++ + F+R+ ++++ + V + S + AD+FTK
Sbjct: 358 NHQVYHERTKHIDIRLHFIRDMIESKEIVVEKMASEENPADVFTK 492
>BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vulgaris},
partial (13%)
Length = 494
Score = 90.1 bits (222), Expect = 6e-18
Identities = 50/125 (40%), Positives = 76/125 (60%), Gaps = 1/125 (0%)
Frame = +1
Query: 1361 RPEISFSVNKVCQFLSQPLEEHWKAVKRILRYLKGTIHHGLHIRPCSLHSPV-SLVAYCD 1419
RP+I +SV+ + +F+ P + H A RILRY++GT+ +GL + P S V L+ Y D
Sbjct: 121 RPDICYSVSVISKFMHDPRKPHLIAANRILRYVRGTMEYGL-LFPYGAKSEVYELICYSD 297
Query: 1420 ADWGSDPDDRRSTSGSCVFFGPNLVTWTSKKQTLVARSSTEAEYRSLANTSAELLWIQSL 1479
+DW D RRSTSG F ++W +KKQ + A SS EAEY + + + LW+ S+
Sbjct: 298 SDWCGD---RRSTSGYVFKFNDAAISWCTKKQPITALSSYEAEYIAGTFATFQALWLDSV 468
Query: 1480 LQELQ 1484
++EL+
Sbjct: 469 IKELK 483
>BG586293 weakly similar to PIR|E84473|E84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 763
Score = 89.7 bits (221), Expect = 8e-18
Identities = 45/89 (50%), Positives = 61/89 (67%)
Frame = +2
Query: 1087 TWTLVTLPKGRHPIGCKWVFRIKENADGSVNRYKARLVAKGYHQVKGFDYAETFSPVVKP 1146
T LV P G PIG +W+++IK N DG++ +YKARLVAKGY + +G D+ E F+PVV+
Sbjct: 56 TLKLVKKPTGVKPIGLRWIYKIKRNEDGTLIKYKARLVAKGYVKQQGIDFDEVFAPVVRI 235
Query: 1147 ITIRLILTLAVSKRWHIHQLDVNNAFLHG 1175
TI L+L LA + IH +DV AFL+G
Sbjct: 236 ETI*LLLALAATNGC*IHHIDVKIAFLNG 322
>BI262917 weakly similar to GP|19920130|g Putative retroelement {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (8%)
Length = 426
Score = 81.6 bits (200), Expect = 2e-15
Identities = 42/110 (38%), Positives = 60/110 (54%)
Frame = +1
Query: 786 HTHHQNGSVERKHRHIVETGLSLLACANMPTSYWDHAFLTATFLINRLPTPVLGNLSPYH 845
HT QNG ER +R ++E ++L A M S+W A TA ++INR P+ V+ +P
Sbjct: 82 HTPQQNGVAERMNRTLLERTRAMLKTAGMAKSFWAEAVKTACYVINRSPSTVIDLKTPME 261
Query: 846 KLFKVPADYKILRVFGSACYPHLRPYNSSKLSFRSQECVFLGYSSSHKGY 895
P DY L VFG Y +KL +S++C+FLGY+ + KGY
Sbjct: 262 MWKGKPVDYSSLHVFGCPVYVMYNSQERTKLDPKSRKCIFLGYADNVKGY 411
>BE941052 weakly similar to PIR|B85188|B85 retrotransposon like protein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 480
Score = 78.6 bits (192), Expect = 2e-14
Identities = 38/73 (52%), Positives = 50/73 (68%)
Frame = +2
Query: 1494 CDNMGAVALTHNPVLHTRTKHMELDIFFVREKVQNQSLFVHHVPSVDQIADIFTKALSPT 1553
CD + A LTHNPV H+R KH+ +DI FVR+ VQ L V HV +VDQ+AD TK LS +
Sbjct: 29 CDYLSATYLTHNPVYHSRMKHISIDIHFVRDLVQQGKLKVQHVCTVDQLADCLTKPLSKS 208
Query: 1554 RFEDLRSKLNVRE 1566
R + LR+K+ V +
Sbjct: 209 RHQLLRNKIGVTD 247
>BI266628
Length = 536
Score = 77.8 bits (190), Expect = 3e-14
Identities = 39/95 (41%), Positives = 59/95 (62%), Gaps = 7/95 (7%)
Frame = -1
Query: 32 LLLKLDDSNF-------LSWKQHVEGIVRTHRLQSFLLHQPEIPPRFLSDADRVNSVENP 84
+++K+D SN L WKQ VEG++R + F++ P+IPP +L+D R + P
Sbjct: 455 VVIKVDPSNKQVLS*VRLQWKQQVEGVLRETKTVKFVI-SPQIPPVYLTDEAREAGTKKP 279
Query: 85 AFLVWEQQDSALFTWLLSSLSPSVLPTVVNCQHSW 119
A+ WE+QDS L TW+LS++ PS+L V +HSW
Sbjct: 278 AYTEWEEQDSLLCTWILSTI*PSLLSRFVLLRHSW 174
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 74.3 bits (181), Expect = 3e-13
Identities = 44/119 (36%), Positives = 66/119 (54%), Gaps = 1/119 (0%)
Frame = +1
Query: 1209 QAPRAWFDRLKAALVKYGFKASCCDPSLFTFHNKS-NVIYILVYVDDIIITGNFLPFIQE 1267
Q+PR WFDR + K+G+ D ++F H+ + ++VYVDDI +TG+ I+
Sbjct: 1 QSPRDWFDRFT*VVKKFGYIQCQTDHAMFIKHSSTVKKAILIVYVDDIFLTGDHGK*IKR 180
Query: 1268 VVQKLNSEFALKQLGQLDYFLGVQVHHLQDRSLLLTQTKYIGDLLERADMAEAKGISTP 1326
+ L EF +K LG L YFLG++V + S ++Q KY+ DLL+ M K I P
Sbjct: 181 LKNLLAEEFEIKDLGNLKYFLGMEVARWKKGS-SISQRKYVLDLLKETRMIGCKTIRDP 354
Score = 35.0 bits (79), Expect = 0.23
Identities = 21/62 (33%), Positives = 31/62 (49%)
Frame = +2
Query: 1314 RADMAEAKGISTPMVSGARLSKHGADYFSDPTLYRSIVGALQYATLTRPEISFSVNKVCQ 1373
R + + K TPM + +L D Y+ +VG L Y + TRP+ISF V + Q
Sbjct: 317 RQE*LDVKPSETPMDATVKLGTLDNGTLVDKGRYQRLVGKLIYLSHTRPDISFVVCTMSQ 496
Query: 1374 FL 1375
F+
Sbjct: 497 FM 502
>CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotiana tabacum},
partial (7%)
Length = 780
Score = 72.8 bits (177), Expect = 1e-12
Identities = 38/90 (42%), Positives = 53/90 (58%)
Frame = -2
Query: 1140 FSPVVKPITIRLILTLAVSKRWHIHQLDVNNAFLHGALQEEVYMQQPAGFQNSDKTLVCK 1199
F P+VK TI +L++ + ++ LDV AFL G L E++YM QP GF +V K
Sbjct: 554 FVPIVKLNTIMFLLSIVAIENLYLE*LDVKTAFLRGDLVEDIYMHQPEGFS*EVGKMVGK 375
Query: 1200 LNKALYGLKQAPRAWFDRLKAALVKYGFKA 1229
L K++YGLKQ PR LKA + GF++
Sbjct: 374 LKKSMYGLKQGPRQCI*SLKALCTRKGFRS 285
>BG582205
Length = 801
Score = 53.9 bits (128), Expect = 5e-07
Identities = 52/164 (31%), Positives = 78/164 (46%), Gaps = 15/164 (9%)
Frame = +3
Query: 652 LTTCKIPVPHKSKYT-ICHSCCVAKSHRLPSSASSTLYTAPLELIFADLWGPASIESSAG 710
LT K+ + ++ K + C CC+ K+HRLPS + P + S
Sbjct: 342 LTVLKLHLVYQIKLSDFCSFCCLGKAHRLPS------------------FHPLYLTLSLL 467
Query: 711 YSYFLTCVDAFSR---------FTWIYLLKRKSETS--TVFIQFQAMVELKFGL--KIKS 757
S+F+ C D SR F++ + K+E S T+F F+ +VEL++ L ++ S
Sbjct: 468 NSFFVICGDYSSRVFFNLCGCLFSF*LDISFKTEVSHFTIFQNFKRIVELQYKLLHQVCS 647
Query: 758 VQTDGGTEFKPLLPHFLKLG-ITHRLTCPHTHHQNGSVERKHRH 800
GG EF+P + + LG H LT + GSVERKHRH
Sbjct: 648 NCLRGG-EFRPFITYLNNLG*PAHTLTT-----KMGSVERKHRH 761
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.133 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 55,243,803
Number of Sequences: 36976
Number of extensions: 960744
Number of successful extensions: 13356
Number of sequences better than 10.0: 423
Number of HSP's better than 10.0 without gapping: 7813
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10902
length of query: 1573
length of database: 9,014,727
effective HSP length: 109
effective length of query: 1464
effective length of database: 4,984,343
effective search space: 7297078152
effective search space used: 7297078152
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0010.11


