

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
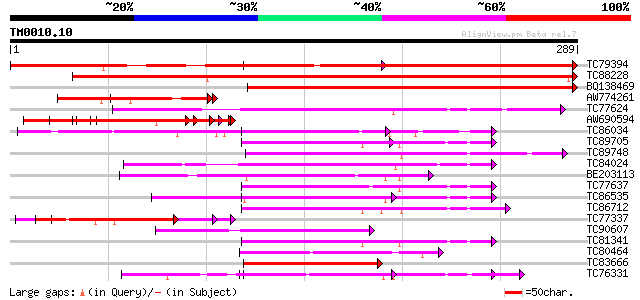
Query= TM0010.10
(289 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC79394 weakly similar to PIR|T49019|T49019 probable RNA binding... 431 e-121
TC88228 similar to GP|13605916|gb|AAK32943.1 AT4g00830/A_TM018A1... 262 1e-70
BQ138469 weakly similar to PIR|T49019|T49 probable RNA binding p... 231 3e-61
AW774261 weakly similar to EGAD|146423|156 vitellogenin {Anolis ... 80 1e-15
TC77624 similar to PIR|T12196|T12196 RNA-binding protein - fava ... 77 6e-15
AW690594 similar to GP|23498163|emb hypothetical protein {Plasmo... 69 2e-12
TC86034 similar to GP|6815067|dbj|BAA90354.1 ESTs AU082210(C5365... 65 3e-11
TC89705 similar to GP|21689785|gb|AAM67536.1 unknown protein {Ar... 60 1e-09
TC89748 homologue to PIR|B84565|B84565 probable spliceosome asso... 59 3e-09
TC84024 similar to PIR|S77714|S77714 RNA-binding protein precurs... 58 4e-09
BE203113 weakly similar to GP|18026763|gb| phragmoplastin-intera... 58 5e-09
TC77637 similar to GP|15292697|gb|AAK92717.1 putative RNA-bindin... 57 6e-09
TC86535 similar to GP|9758076|dbj|BAB08520.1 contains similarity... 56 1e-08
TC86712 similar to GP|17528972|gb|AAL38696.1 putative RNA-bindin... 55 2e-08
TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Atel... 53 2e-07
TC90607 similar to GP|20466724|gb|AAM20679.1 unknown protein {Ar... 52 2e-07
TC81341 similar to GP|15292697|gb|AAK92717.1 putative RNA-bindin... 52 2e-07
TC80464 weakly similar to GP|13548328|emb|CAC35875. putative pro... 52 3e-07
TC83666 similar to GP|15042832|gb|AAK82455.1 putative RNA bindin... 51 5e-07
TC76331 similar to GP|7673355|gb|AAF66823.1| poly(A)-binding pro... 51 5e-07
>TC79394 weakly similar to PIR|T49019|T49019 probable RNA binding protein -
Arabidopsis thaliana, partial (74%)
Length = 1909
Score = 431 bits (1109), Expect = e-121
Identities = 223/293 (76%), Positives = 244/293 (83%), Gaps = 4/293 (1%)
Frame = +2
Query: 1 MHKMPGTRQRDVTAQNPESPEGNSEPEKPTDSDEQVDFDGDNDQEE----EIEYEEVEEE 56
+H+MPGTRQ+D TA+N ESP GNSEPEKPTDS EQVD DGDNDQEE E+EYEEVE E
Sbjct: 131 IHRMPGTRQKDATARNSESPGGNSEPEKPTDSGEQVDLDGDNDQEESSEEEVEYEEVEVE 310
Query: 57 EEVEDEEVEEVEEEEEEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEKKKHAELLALPPH 116
EEVE EEEEEEEEEEVEEESKP D E DE +KKKHAELLALPPH
Sbjct: 311 EEVE---------EEEEEEEEEEVEEESKPLDEE---------DEADKKKHAELLALPPH 436
Query: 117 GSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAI 176
GSEVYIGGI + SE+DLRVFCQSVGEV+EVR+MK KE+ KGYAFV FKTKELAS A+
Sbjct: 437 GSEVYIGGIPHETSEKDLRVFCQSVGEVAEVRVMKGKEA---KGYAFVTFKTKELASNAL 607
Query: 177 EELNNSEFKGKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQS 236
+ELNNSEFKG+KIKCS SQ KHRLFIG++PK+WTVEDMKKVVA VGPGVIS+ELLKD QS
Sbjct: 608 KELNNSEFKGRKIKCSPSQVKHRLFIGSVPKEWTVEDMKKVVAKVGPGVISVELLKDPQS 787
Query: 237 SGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWADPRNSESSAVS 289
S RNRGFAFIEY+NHACAEYSRQKMSNSNFKL+NN VSWADPRNSESS+ S
Sbjct: 788 SSRNRGFAFIEYHNHACAEYSRQKMSNSNFKLDNNDAIVSWADPRNSESSSSS 946
Score = 42.4 bits (98), Expect = 2e-04
Identities = 19/73 (26%), Positives = 38/73 (52%)
Frame = +2
Query: 120 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEEL 179
VY+ + N+++ L+ + G++++V + AK E Y FV F + A +A++
Sbjct: 959 VYVKNLPENITQNRLKELFEHHGKITKVALPPAKAGQEKSRYGFVHFADRSSAMKALKNT 1138
Query: 180 NNSEFKGKKIKCS 192
E G+ ++CS
Sbjct: 1139 EKYEINGQTLECS 1177
>TC88228 similar to GP|13605916|gb|AAK32943.1 AT4g00830/A_TM018A10_14
{Arabidopsis thaliana}, partial (67%)
Length = 1712
Score = 262 bits (670), Expect = 1e-70
Identities = 130/270 (48%), Positives = 186/270 (68%), Gaps = 13/270 (4%)
Frame = +1
Query: 33 DEQVDFDGDNDQEE-EIEYEEVEEEEEVEDEEVEEVEEEEEEEEEEEEVEEESKPSDGEE 91
D++VD D +N EE + + EE +++ V+ E E E EE E EE E + E
Sbjct: 175 DDRVDLDEENYMEEMDDDVEEQIDDDGVDGGEDENAEGSVEEHEYEETAAEAGQKDQFPE 354
Query: 92 EMRVDHTN----------DEVEKKKHAELLALPPHGSEVYIGGIANNVSEEDLRVFCQSV 141
+ DH DE E++KH ELL+ PPHGSEV+IGG+ + S++D+R C+ +
Sbjct: 355 GEKSDHGAEEDELKPALIDEEEREKHDELLSRPPHGSEVFIGGLPRDTSDDDVRELCEPM 534
Query: 142 GEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEELNNSEFKGKKIKCSSSQAKHRLF 201
G++ E++++K +E+GE KGYAFV +KTKE+A +AI++++N EFKGK ++C S+ KHRLF
Sbjct: 535 GDIVEIKLIKDRETGESKGYAFVGYKTKEVAQKAIDDIHNKEFKGKTLRCLLSETKHRLF 714
Query: 202 IGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKM 261
IGNIPK WT ++ +K V VGPGV SI+L+KD Q+ RNRGFAF+ YYN+ACA++SRQKM
Sbjct: 715 IGNIPKTWTEDEFRKAVEGVGPGVESIDLIKDPQNQSRNRGFAFVLYYNNACADFSRQKM 894
Query: 262 SNSNFKLENNAPTVSWADPRNS--ESSAVS 289
S+ FKL+ PTV+WADP+ S +S+A S
Sbjct: 895 SSVGFKLDGITPTVTWADPKTSPDQSAAAS 984
>BQ138469 weakly similar to PIR|T49019|T49 probable RNA binding protein -
Arabidopsis thaliana, partial (42%)
Length = 633
Score = 231 bits (588), Expect = 3e-61
Identities = 105/168 (62%), Positives = 140/168 (82%)
Frame = +2
Query: 122 IGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEELNN 181
+GGI + EDL+ FC+ +GEV +VRIMK K++ E KG+AFV ++ ELAS+AIEELNN
Sbjct: 2 VGGIPLDAKSEDLKEFCERIGEVVQVRIMKGKDASENKGFAFVTYRNVELASKAIEELNN 181
Query: 182 SEFKGKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNR 241
+EFKG+KIKCS+SQAK+RLFIGNIP+ W +D+KKVV D+GPGV ++EL+KD+++ NR
Sbjct: 182 TEFKGRKIKCSTSQAKNRLFIGNIPRSWGEKDLKKVVTDIGPGVTAVELVKDMKNISNNR 361
Query: 242 GFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWADPRNSESSAVS 289
GFAFI+Y+N+ CAEY RQKM + +FKL +N+PTVSWADP+NS+SSA S
Sbjct: 362 GFAFIDYHNNQCAEYGRQKMMSPSFKLGDNSPTVSWADPKNSDSSASS 505
>AW774261 weakly similar to EGAD|146423|156 vitellogenin {Anolis pulchellus},
partial (40%)
Length = 495
Score = 79.7 bits (195), Expect = 1e-15
Identities = 47/84 (55%), Positives = 60/84 (70%), Gaps = 5/84 (5%)
Frame = +1
Query: 25 EPEKPTDSDEQVDFDGDNDQE---EEIEYEEVEEEEEVE--DEEVEEVEEEEEEEEEEEE 79
EPEK +SDE++D + +ND E EEIEYEEVEEEEEVE +EEVEE EE+ EEEEEE E
Sbjct: 256 EPEKTVESDEKIDLEEENDPEEEMEEIEYEEVEEEEEVEEIEEEVEENEEDAEEEEEE*E 435
Query: 80 VEEESKPSDGEEEMRVDHTNDEVE 103
EEE + EE+ + + +D+ E
Sbjct: 436 KEEE----EVEEDDTMQNLDDDDE 495
Score = 41.6 bits (96), Expect = 4e-04
Identities = 23/55 (41%), Positives = 34/55 (61%)
Frame = +1
Query: 52 EVEEEEEVEDEEVEEVEEEEEEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEKKK 106
E E E+ VE +E ++EEE + EEE EE+E E + EEE V+ +EVE+ +
Sbjct: 250 EPEPEKTVESDEKIDLEEENDPEEEMEEIEYE----EVEEEEEVEEIEEEVEENE 402
>TC77624 similar to PIR|T12196|T12196 RNA-binding protein - fava bean
(fragment), partial (90%)
Length = 1214
Score = 77.4 bits (189), Expect = 6e-15
Identities = 62/242 (25%), Positives = 106/242 (43%), Gaps = 11/242 (4%)
Frame = +1
Query: 53 VEEEEEVEDEEVEEVEEEEEEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEKKKHAELLA 112
V +EE V E EE EEEE+EEEEV EE + GE VD
Sbjct: 253 VAQEEAVVAVEDEEKVVVEEEEKEEEEVVEEKENIGGETVAEVDTR-------------- 390
Query: 113 LPPHGSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELA 172
+++Y G + +V L + G + ++ +++G+ +G+AFV E
Sbjct: 391 -----TKLYFGNLPYSVDSALLAGLIEEYGSAELIEVLYDRDTGKSRGFAFVTMSCVEDC 555
Query: 173 SQAIEELNNSEFKGKKIKCSSS-----------QAKHRLFIGNIPKKWTVEDMKKVVADV 221
+ I+ L+ EF G+ ++ + S + +++LF+GN+ T E + + +
Sbjct: 556 NAVIQNLDGKEFMGRTLRVNFSDKPKPKEPLYPETEYKLFVGNLAWTVTSESLTQAFQEH 735
Query: 222 GPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWADPR 281
G V+ +L D +G++RG+ F+ Y +E N +LE VS A +
Sbjct: 736 GT-VVGARVLFD-GETGKSRGYGFVSYATK--SEMDTALAIMDNVELEGRTLRVSLAQGK 903
Query: 282 NS 283
S
Sbjct: 904 RS 909
>AW690594 similar to GP|23498163|emb hypothetical protein {Plasmodium
falciparum 3D7}, partial (10%)
Length = 633
Score = 68.9 bits (167), Expect = 2e-12
Identities = 40/85 (47%), Positives = 52/85 (61%)
Frame = +3
Query: 8 RQRDVTAQNPESPEGNSEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEV 67
++R V N E EG E E + ++D+EEE E++E EEEEE E+EE E
Sbjct: 30 KRRKVEEHNEEEDEGEVEEE-----------EDEHDEEEEDEHDEEEEEEEEEEEEDEHD 176
Query: 68 EEEEEEEEEEEEVEEESKPSDGEEE 92
+EEEEEEEEEEE EEE +GEE+
Sbjct: 177 DEEEEEEEEEEE-EEEDDDEEGEED 248
Score = 60.8 bits (146), Expect = 6e-10
Identities = 34/80 (42%), Positives = 49/80 (60%)
Frame = +3
Query: 35 QVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEEEEEVEEESKPSDGEEEMR 94
+V+ + + E E+E EE E +EE EDE EE EEEEEEEEE+E +EE + + EEE
Sbjct: 39 KVEEHNEEEDEGEVEEEEDEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEEEEEEEEEEEE 218
Query: 95 VDHTNDEVEKKKHAELLALP 114
D ++E E+ + + LP
Sbjct: 219 ED-DDEEGEEDEIDRISTLP 275
Score = 58.2 bits (139), Expect = 4e-09
Identities = 27/74 (36%), Positives = 47/74 (63%)
Frame = +3
Query: 42 NDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEEEEEVEEESKPSDGEEEMRVDHTNDE 101
N++E+E E EE E+E + E+E+ + EEEEEEEEEEE+ ++ + + EEE + +DE
Sbjct: 54 NEEEDEGEVEEEEDEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEEEEEEEEEEEEEDDDE 233
Query: 102 VEKKKHAELLALPP 115
++ + ++ P
Sbjct: 234 EGEEDEIDRISTLP 275
Score = 57.8 bits (138), Expect = 5e-09
Identities = 27/72 (37%), Positives = 47/72 (64%)
Frame = +3
Query: 33 DEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEEEEEVEEESKPSDGEEE 92
+E + + + + EEE + + EEE+E ++EE EE EEEEE+E ++EE EEE + + EE+
Sbjct: 45 EEHNEEEDEGEVEEEEDEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEEEEEEEEEEEEED 224
Query: 93 MRVDHTNDEVEK 104
+ DE+++
Sbjct: 225 DDEEGEEDEIDR 260
Score = 56.6 bits (135), Expect = 1e-08
Identities = 33/78 (42%), Positives = 49/78 (62%), Gaps = 2/78 (2%)
Frame = +3
Query: 21 EGNSEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEE--EEEEE 78
E + + K + +E+ D +G+ ++EE+ EE E+E + E+EE EE EEE+E EEEEE
Sbjct: 18 ESSWKRRKVEEHNEEED-EGEVEEEEDEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEEEE 194
Query: 79 EVEEESKPSDGEEEMRVD 96
E EEE + D +EE D
Sbjct: 195EEEEEEEEEDDDEEGEED 248
Score = 50.4 bits (119), Expect = 8e-07
Identities = 28/65 (43%), Positives = 39/65 (59%)
Frame = +3
Query: 45 EEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEK 104
E + +VEE E EDE EVEEEE+E +EEEE E + + + EEE D +DE E+
Sbjct: 18 ESSWKRRKVEEHNEEEDEG--EVEEEEDEHDEEEEDEHDEEEEEEEEEEEEDEHDDEEEE 191
Query: 105 KKHAE 109
++ E
Sbjct: 192 EEEEE 206
>TC86034 similar to GP|6815067|dbj|BAA90354.1 ESTs AU082210(C53655)
AU056546(S20671) AU056545(S20671) correspond to a
region of the predicted, partial (42%)
Length = 2412
Score = 65.1 bits (157), Expect = 3e-11
Identities = 63/302 (20%), Positives = 129/302 (41%), Gaps = 58/302 (19%)
Frame = +3
Query: 5 PGTRQRDVTAQNPESPEGNSEPEKPTDSDE-QVDFDGDNDQEEEIEYEEVEEEEEVEDEE 63
P +Q +V + P + E E P + D+ Q + +G+ + EEE+ EE E++++ EDE
Sbjct: 156 PEPQQEEVAEEEPTTQE---EENTPIEEDQPQNEGEGEEEPEEEVAEEE-EDDQQGEDET 323
Query: 64 VEEVEEEEEEEEEEEEVEEES----------KPSDGEEEMRVDHTNDEVEK--------K 105
+E ++++ E E + ++GEEE ++ ++ VEK +
Sbjct: 324 LENQQQQDGGEASNNAAVSEQTVTVEANGGGENNNGEEEEDLELEDEPVEKLLEPFTKEQ 503
Query: 106 KHA-----------------ELLALPPHGSEVYIGGIANNVSEEDLRVFCQSVGEVSEVR 148
HA +L + P ++++ G+ + + E L GE+ + +
Sbjct: 504 LHALVKQAVEKYPDLSENVRQLADVDPAHRKIFVHGLGWDATAETLTTTFSKYGEIEDCK 683
Query: 149 IMKAKESGEGKGYAFVAFKTKELASQAIEELNNSEFKGKKIKCS---------------- 192
+ K SG+ KGYAF+ FK + +A+++ + C
Sbjct: 684 AVTDKASGKSKGYAFILFKHRTGCRRALKQ-PQKVIGNRTTSCQLASAGPVPAPPPNAPP 860
Query: 193 -SSQAKHRLFIGNI-----PKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFI 246
S + ++F+ N+ P+K + + K +V G + ++ +++G+ +GFA
Sbjct: 861 VSEYTQRKIFVSNVSSDIDPQK--LLEFFKQFGEVDDGPLGLD-----KATGKPKGFALF 1019
Query: 247 EY 248
Y
Sbjct: 1020VY 1025
Score = 44.3 bits (103), Expect = 6e-05
Identities = 21/76 (27%), Positives = 43/76 (55%)
Frame = +3
Query: 119 EVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEE 178
++++ +++++ + L F + GEV + + K +G+ KG+A +KT E A +A+EE
Sbjct: 882 KIFVSNVSSDIDPQKLLEFFKQFGEVDDGPLGLDKATGKPKGFALFVYKTVESAKKALEE 1061
Query: 179 LNNSEFKGKKIKCSSS 194
N F+G + C +
Sbjct: 1062-PNKVFEGHTLYCQKA 1106
>TC89705 similar to GP|21689785|gb|AAM67536.1 unknown protein {Arabidopsis
thaliana}, partial (46%)
Length = 812
Score = 60.1 bits (144), Expect = 1e-09
Identities = 35/138 (25%), Positives = 78/138 (56%), Gaps = 8/138 (5%)
Frame = +3
Query: 119 EVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEE 178
+++IGG+A + + E+ + + GE+++ IMK + +G +G+ F+ + + Q I+E
Sbjct: 114 KIFIGGLAKDTTLEEFVKYFERYGEITDSVIMKDRHTGRPRGFGFITYADASVVDQVIQE 293
Query: 179 ---LNNSEFKGKK-IKCSSSQAK----HRLFIGNIPKKWTVEDMKKVVADVGPGVISIEL 230
+N+ + + K+ I +SQ ++F+G IP + +++K + G V+ E+
Sbjct: 294 NHIINDKQVEIKRTIPKGASQTNDFKTKKIFVGGIPATVSEDELKIFFSKHG-NVVEHEI 470
Query: 231 LKDLQSSGRNRGFAFIEY 248
++D ++ R+RGF F+ +
Sbjct: 471 IRD-HTTKRSRGFGFVVF 521
Score = 42.4 bits (98), Expect = 2e-04
Identities = 21/78 (26%), Positives = 41/78 (51%)
Frame = +3
Query: 119 EVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEE 178
++++GGI VSE++L++F G V E I++ + +G+ FV F + + + +
Sbjct: 378 KIFVGGIPATVSEDELKIFFSKHGNVVEHEIIRDHTTKRSRGFGFVVFDSDKAVDNLLAD 557
Query: 179 LNNSEFKGKKIKCSSSQA 196
N + +I C +S A
Sbjct: 558 GNMIDMDDTQI-CGTSHA 608
>TC89748 homologue to PIR|B84565|B84565 probable spliceosome associated
protein [imported] - Arabidopsis thaliana, partial (56%)
Length = 707
Score = 58.5 bits (140), Expect = 3e-09
Identities = 48/171 (28%), Positives = 86/171 (50%), Gaps = 7/171 (4%)
Frame = +2
Query: 121 YIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEELN 180
Y+G + +SEE L G V V + K + + + +GY FV F+++E A AI+ LN
Sbjct: 173 YVGNLDPQISEELLWELFVQAGPVVNVYVPKDRVTNQHQGYGFVEFRSEEDADYAIKVLN 352
Query: 181 NSEFKGKKIKCS-SSQAKH------RLFIGNIPKKWTVEDMKKVVADVGPGVISIELLKD 233
+ GK I+ + +SQ K LFIGN+ + + + G V + ++++D
Sbjct: 353 MIKLYGKPIRVNKASQDKKSLDVGANLFIGNLDPDVDEKLLYDTFSAFGVIVTNPKIMRD 532
Query: 234 LQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWADPRNSE 284
+ +RGF FI Y + ++ + + M+ L N TVS+A ++++
Sbjct: 533 -PDTRNSRGFGFISYDSFEASDSAIEAMNGQ--YLCNRQITVSYAYKKDTK 676
>TC84024 similar to PIR|S77714|S77714 RNA-binding protein precursor 33K -
wood tobacco, partial (55%)
Length = 867
Score = 58.2 bits (139), Expect = 4e-09
Identities = 44/213 (20%), Positives = 91/213 (42%), Gaps = 23/213 (10%)
Frame = +3
Query: 59 VEDEEVEEVEEEEEEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEKKKHAELLALPPHGS 118
V +E E ++ +++E E E KP +EE +V ++D
Sbjct: 207 VATDEFETSQDTIISQQQEPEPETSDKPKQ-DEEQKVTTSDD----------------AG 335
Query: 119 EVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEE 178
+Y+G + +++ L G V V I+ + + +G+AFV K+ E A +AI
Sbjct: 336 RLYVGNLPFSMTSSQLADIFAEAGTVVSVEIVYDRLTDRSRGFAFVTMKSAEEAKEAIRM 515
Query: 179 LNNSEFKGKKIKCSSSQ-----------------------AKHRLFIGNIPKKWTVEDMK 215
+ S+ G+ K + + + H+++ GN+ +D++
Sbjct: 516 FDGSQVGGRSAKVNFPEVPKGGERLVMGQKVRNSYRGFVDSPHKIYAGNLGWGMNSQDLR 695
Query: 216 KVVADVGPGVISIELLKDLQSSGRNRGFAFIEY 248
D PG++ +++ + + +GR+RGF F+ +
Sbjct: 696 DAF-DEQPGLLGAKIIYE-RDNGRSRGFGFVTF 788
>BE203113 weakly similar to GP|18026763|gb| phragmoplastin-interacting
protein PHIP1 {Arabidopsis thaliana}, partial (11%)
Length = 548
Score = 57.8 bits (138), Expect = 5e-09
Identities = 41/181 (22%), Positives = 91/181 (49%), Gaps = 21/181 (11%)
Frame = +1
Query: 57 EEVEDEEVEEVEEEEEEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEKKKHAELLALPPH 116
+ + E+ ++ + +++ ++ + EE K EE + +N + E + EL
Sbjct: 1 KSMNKEKQKKKNQLKKKRKKNKTAEENGKAKIAEE----NDSNHQEEMPQSVELANTTTT 168
Query: 117 GSE--------VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKT 168
S+ VY+GGI SE+D+ + + G ++E+ M ++G+ +G A ++FKT
Sbjct: 169 TSQENGDVATKVYVGGIPYYSSEDDIHSYFEGCGTITEINCMTFPDTGKFRGIAIISFKT 348
Query: 169 KELASQAIEELNNSEFKGKKIK---CSSSQAK----------HRLFIGNIPKKWTVEDMK 215
+ A +A+ L+ ++ G +K C ++QA +R+++G++ + T E+++
Sbjct: 349 EAAAKRAL-ALDGADMGGLFLKIQPCKATQATRFTPEMKEGYNRIYVGSLSWEITEEELR 525
Query: 216 K 216
K
Sbjct: 526 K 528
>TC77637 similar to GP|15292697|gb|AAK92717.1 putative RNA-binding protein
{Arabidopsis thaliana}, partial (59%)
Length = 1724
Score = 57.4 bits (137), Expect = 6e-09
Identities = 39/152 (25%), Positives = 71/152 (46%), Gaps = 22/152 (14%)
Frame = +2
Query: 119 EVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEE 178
+++IGGI+ + +EE LR + + GEV E IMK + +G +G+ FV F +A ++E
Sbjct: 389 KLFIGGISWDTNEERLREYFSTYGEVKEAVIMKDRTTGRARGFGFVVFIDPAVADIVVQE 568
Query: 179 LNNSEFKGKKIKCSSSQAK----------------------HRLFIGNIPKKWTVEDMKK 216
+N + G+ ++ + + ++F+G + T D KK
Sbjct: 569 KHNID--GRMVEAKKAVPRDDQNVLSRTSGSIHGSPGPGRTRKIFVGGLASTVTESDFKK 742
Query: 217 VVADVGPGVISIELLKDLQSSGRNRGFAFIEY 248
G + + ++ D ++ R RGF FI Y
Sbjct: 743 YFDQFGT-ITDVVVMYD-HNTQRPRGFGFITY 832
>TC86535 similar to GP|9758076|dbj|BAB08520.1 contains similarity to RNA
binding protein~gene_id:MNF13.1 {Arabidopsis thaliana},
partial (48%)
Length = 1555
Score = 56.2 bits (134), Expect = 1e-08
Identities = 24/79 (30%), Positives = 45/79 (56%)
Frame = +1
Query: 119 EVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEE 178
++++GGI +NV+E++ R F GEV + +IM+ + +G+ FV F T+E +
Sbjct: 535 KIFVGGIPSNVTEDEFRDFFTRYGEVKDHQIMRDHSTNRSRGFGFVTFDTEEAVDDLLSM 714
Query: 179 LNNSEFKGKKIKCSSSQAK 197
N EF G +++ ++ K
Sbjct: 715 GNKIEFAGTQVEIKKAEPK 771
Score = 55.8 bits (133), Expect = 2e-08
Identities = 40/187 (21%), Positives = 85/187 (45%), Gaps = 11/187 (5%)
Frame = +1
Query: 73 EEEEEEEVEEESKPSDGEEEMRVDHTNDEVEKKKHAELLALPPHGS---EVYIGGIANNV 129
+ + E + E G E+ V + +++H +L G+ +++IGG+A
Sbjct: 124 QNQSMESPKNEHIIGTGGEQNDVGRNYEHEHEQQHHNSQSLTGDGASPGKIFIGGLARET 303
Query: 130 SEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEELNNSEFKGKKI 189
+ GE+++ IMK +++G+ +G+ F+ + + + IE+ + K +I
Sbjct: 304 TIAQFIKHFGKYGEITDSVIMKDRKTGQPRGFGFITYADPSVVDKVIEDSHIINSKQVEI 483
Query: 190 K--------CSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNR 241
K S ++F+G IP T ++ + G V ++++D S+ R+R
Sbjct: 484 KRTIPRGAVGSKDFRTKKIFVGGIPSNVTEDEFRDFFTRYGE-VKDHQIMRD-HSTNRSR 657
Query: 242 GFAFIEY 248
GF F+ +
Sbjct: 658 GFGFVTF 678
>TC86712 similar to GP|17528972|gb|AAL38696.1 putative RNA-binding protein
{Arabidopsis thaliana}, partial (51%)
Length = 2144
Score = 55.5 bits (132), Expect = 2e-08
Identities = 40/159 (25%), Positives = 74/159 (46%), Gaps = 22/159 (13%)
Frame = +3
Query: 119 EVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEE 178
+++IGGI+ + +E+ LR + Q+ G+V E IMK + +G +G+ FV F +A + + E
Sbjct: 186 KLFIGGISWDTNEDRLRQYFQNFGDVVEAVIMKDRTTGRARGFGFVVFADPSVAERVVME 365
Query: 179 ---LNNSEFKGKK--------IKCSSSQAKH-----------RLFIGNIPKKWTVEDMKK 216
++ + KK + S+ + H ++F+G + T D K
Sbjct: 366 KHVIDGRTVEAKKAVPRDDQNVFTRSNSSSHGSPAPTPIRTKKIFVGGLASTVTESDFKN 545
Query: 217 VVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAE 255
G + + ++ D ++ R RGF FI Y + E
Sbjct: 546 YFDQFGT-ITDVVVMYD-HNTQRPRGFGFITYDSEEAVE 656
>TC77337 weakly similar to GP|4019275|gb|AAC95573.1| orf 48 {Ateline
herpesvirus 3}, partial (8%)
Length = 986
Score = 52.8 bits (125), Expect = 2e-07
Identities = 29/74 (39%), Positives = 49/74 (66%), Gaps = 1/74 (1%)
Frame = +3
Query: 14 AQNPESPEGNSEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEE-VEEEEE 72
+ + ++PEG + + +E+ D DGD+ E++ E E+ ++EEE +E+ EE V+EE+
Sbjct: 540 SNSKKAPEGGAGGADE-NGEEEDDEDGDDQDEDDDEDEDDDDEEEGGEEDEEEGVDEEDN 716
Query: 73 EEEEEEEVEEESKP 86
EEEEE+E EE +P
Sbjct: 717 EEEEEDEDEEALQP 758
Score = 43.9 bits (102), Expect = 7e-05
Identities = 35/139 (25%), Positives = 63/139 (45%), Gaps = 27/139 (19%)
Frame = +3
Query: 4 MPGTRQRDVTAQNPESPEGNSEPEKPTDSDEQVDFDGDN---DQEEEIEYEE-------- 52
MP T+ T + + ++E E D +++ D DGD + E+E+ E+
Sbjct: 357 MPTTKGDSKTQS--QDKQHDAEGEDGNDDEDEEDDDGDGAFGEGEDELSSEDGGGYGNNS 530
Query: 53 ----------------VEEEEEVEDEEVEEVEEEEEEEEEEEEVEEESKPSDGEEEMRVD 96
+E E ED+E + ++E+++E+E+++ EEE D EE VD
Sbjct: 531 NNKSNSKKAPEGGAGGADENGEEEDDEDGDDQDEDDDEDEDDDDEEEGGEED--EEEGVD 704
Query: 97 HTNDEVEKKKHAELLALPP 115
++E E++ E PP
Sbjct: 705 EEDNEEEEEDEDEEALQPP 761
Score = 40.4 bits (93), Expect = 8e-04
Identities = 23/85 (27%), Positives = 43/85 (50%)
Frame = +3
Query: 22 GNSEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEEEEEVE 81
GN+ K G D+ E E +E ++++ +D+E E+ ++EEE EE+EE
Sbjct: 519 GNNSNNKSNSKKAPEGGAGGADENGEEEDDEDGDDQDEDDDEDEDDDDEEEGGEEDEEEG 698
Query: 82 EESKPSDGEEEMRVDHTNDEVEKKK 106
+ + ++ EEE + +K+K
Sbjct: 699 VDEEDNEEEEEDEDEEALQPPKKRK 773
Score = 35.4 bits (80), Expect = 0.026
Identities = 20/64 (31%), Positives = 34/64 (52%), Gaps = 3/64 (4%)
Frame = +3
Query: 15 QNPESPEGNSEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEE---EEVEDEEVEEVEEEE 71
+N E + ++ D DE D D + + EE E E V+EE EE EDE+ E ++ +
Sbjct: 585 ENGEEEDDEDGDDQDEDDDEDEDDDDEEEGGEEDEEEGVDEEDNEEEEEDEDEEALQPPK 764
Query: 72 EEEE 75
+ ++
Sbjct: 765 KRKK 776
>TC90607 similar to GP|20466724|gb|AAM20679.1 unknown protein {Arabidopsis
thaliana}, partial (49%)
Length = 1143
Score = 52.4 bits (124), Expect = 2e-07
Identities = 30/112 (26%), Positives = 57/112 (50%)
Frame = +1
Query: 75 EEEEEVEEESKPSDGEEEMRVDHTNDEVEKKKHAELLALPPHGSEVYIGGIANNVSEEDL 134
+ ++ E E+K +G + + E + A + ++++ G++ SE+ L
Sbjct: 463 QPDDNFESENKDYEGRNLENSLNMPNSSEASQEASVKT-----KKLFVTGLSFYTSEKTL 627
Query: 135 RVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEELNNSEFKG 186
R + GE+ EV+++ K S KGYAF+ + T+E AS A++E+N G
Sbjct: 628 RAAFEGFGELVEVKVIIDKISKRSKGYAFIEYTTEEAASAALKEMNGKIING 783
>TC81341 similar to GP|15292697|gb|AAK92717.1 putative RNA-binding protein
{Arabidopsis thaliana}, partial (37%)
Length = 953
Score = 52.4 bits (124), Expect = 2e-07
Identities = 38/150 (25%), Positives = 67/150 (44%), Gaps = 20/150 (13%)
Frame = +2
Query: 119 EVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEE 178
+++IGGI+ + EE L+ + + GEV E IM+ + +G +G+ FV F +A + I +
Sbjct: 278 KLFIGGISWDTDEERLKEYFATYGEVIEAVIMRDRATGRARGFGFVVFSDPAVAERVIID 457
Query: 179 ---LNNSEFKGKKIKCSSSQAK-----------------HRLFIGNIPKKWTVEDMKKVV 218
++ + KK Q ++F+G +P T D K
Sbjct: 458 KHIIDGRTVEAKKAVPRDDQQNINRQTGSVQGSPGPGRTKKIFVGGLPSTITESDFKMYF 637
Query: 219 ADVGPGVISIELLKDLQSSGRNRGFAFIEY 248
G + + ++ D ++ R RGF FI Y
Sbjct: 638 DQFGT-ITDVVVMYD-HNTQRPRGFGFITY 721
>TC80464 weakly similar to GP|13548328|emb|CAC35875. putative protein
{Arabidopsis thaliana}, partial (19%)
Length = 1437
Score = 52.0 bits (123), Expect = 3e-07
Identities = 31/108 (28%), Positives = 58/108 (53%), Gaps = 4/108 (3%)
Frame = +3
Query: 118 SEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIE 177
S + + + V+E+ L+ GE+++V++M+ K +G+ + +AF+ F+T + A AI+
Sbjct: 87 SRICVKNLPKYVAEDRLKQEFSRKGEITDVKLMRTK-NGKSRQFAFIGFRTDQEAQAAIK 263
Query: 178 ELNNSEFKGKKIKCSSSQAKHRLFIGNIPKKW----TVEDMKKVVADV 221
N S + I C +Q +L N+P+ W T +D K + DV
Sbjct: 264 YFNKSYLDTQAITCEVAQ---KLGDANLPRPWSRHSTKKDDKVITTDV 398
Score = 36.6 bits (83), Expect = 0.012
Identities = 61/277 (22%), Positives = 111/277 (40%), Gaps = 10/277 (3%)
Frame = +3
Query: 1 MHKMPGTRQRDVTAQNPESPEGNSEPEK------PTDSDEQVDFDGDN---DQEEEIEYE 51
M K+ V+ P N E E P SD VD +N D+ +I+++
Sbjct: 495 MSKLRANDTSVVSNDGNNQPTLNKETESTLIANHPILSDSHVDELPNNPKSDKSRDIKHD 674
Query: 52 EVEEEEEVEDEEVEEVEEEEEEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEKKKHAELL 111
V + D +V E + E ++ +S SD +++ + H + E +
Sbjct: 675 GVISDM---DYFKSKVTTEWSDSESSDDDSSDSASSDDDDKDKHSHATEHEENR------ 827
Query: 112 ALPPHGSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKEL 171
NN SE R Q + ++ +++ GK A K++
Sbjct: 828 --------------CNNPSETTPRSGAQELD-------LEDQKNTVGKDVA--NDKSQVN 938
Query: 172 ASQAIEELNNSEFK-GKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIEL 230
A++ +L+N E K G C RLF+ N+P T E++++ + G V L
Sbjct: 939 ATEEEGQLSNPEDKKGVSEPC-------RLFVRNLPYSTTEEELEEHFSQFG-SVSQAHL 1094
Query: 231 LKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFK 267
+ D + + R++G A+I + A + ++ NS F+
Sbjct: 1095VVD-KDTKRSKGIAYIHFSAPEFAARALEESDNSIFQ 1202
>TC83666 similar to GP|15042832|gb|AAK82455.1 putative RNA binding protein
{Oryza sativa}, partial (52%)
Length = 896
Score = 51.2 bits (121), Expect = 5e-07
Identities = 21/71 (29%), Positives = 45/71 (62%)
Frame = +1
Query: 120 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEEL 179
V++GGI + +E D+ GEV ++ +++ K +G+ KG+AF+A++ + + A++ L
Sbjct: 274 VFVGGIPFDFTEGDVIAVFAQYGEVVDINLVRDKGTGKSKGFAFIAYEDQRSTNLAVDNL 453
Query: 180 NNSEFKGKKIK 190
N ++ G+ I+
Sbjct: 454 NGAQVSGRIIR 486
Score = 38.1 bits (87), Expect = 0.004
Identities = 25/77 (32%), Positives = 45/77 (57%), Gaps = 4/77 (5%)
Frame = +1
Query: 176 IEELNNSEFKGKKIKCSSSQAKHR----LFIGNIPKKWTVEDMKKVVADVGPGVISIELL 231
I+++N+ E + +S K++ +F+G IP +T D+ V A G V+ I L+
Sbjct: 190 IQQINSREAALNISEDASWHTKYKDSAYVFVGGIPFDFTEGDVIAVFAQYGE-VVDINLV 366
Query: 232 KDLQSSGRNRGFAFIEY 248
+D + +G+++GFAFI Y
Sbjct: 367 RD-KGTGKSKGFAFIAY 414
>TC76331 similar to GP|7673355|gb|AAF66823.1| poly(A)-binding protein
{Nicotiana tabacum}, partial (97%)
Length = 2581
Score = 51.2 bits (121), Expect = 5e-07
Identities = 36/142 (25%), Positives = 72/142 (50%), Gaps = 2/142 (1%)
Frame = +3
Query: 58 EVEDEEVEEVEEEEEEEEEEEE--VEEESKPSDGEEEMRVDHTNDEVEKKKHAELLALPP 115
E D+ VE ++ +++E V + K S+ E E+++ E K+ A+
Sbjct: 1125 ESTDDAARAVEALNGKKIDDKEWYVGKAQKKSEREHELKIKF---EQSMKEAADKY---- 1283
Query: 116 HGSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQA 175
G+ +Y+ + +++++E L+ S G ++ ++M+ +G +G FVAF T E AS+A
Sbjct: 1284 QGANLYVKNLDDSIADEKLKELFSSYGTITSCKVMR-DPNGVSRGSGFVAFSTPEEASRA 1460
Query: 176 IEELNNSEFKGKKIKCSSSQAK 197
+ E+N K + + +Q K
Sbjct: 1461 LLEMNGKMVASKPLYVTLAQRK 1526
Score = 50.8 bits (120), Expect = 6e-07
Identities = 34/141 (24%), Positives = 69/141 (48%), Gaps = 12/141 (8%)
Frame = +3
Query: 120 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEEL 179
++I + + + L S G + ++ SG+ KGY FV F T+E A +AIE+L
Sbjct: 714 IFIKNLDKAIDHKALHDTFSSFGNILSCKVA-VDGSGQSKGYGFVQFDTEEAAQKAIEKL 890
Query: 180 NNSEFKGKKI-----------KCSSSQAK-HRLFIGNIPKKWTVEDMKKVVADVGPGVIS 227
N K++ + + +AK + +F+ N+ + T +++KK + G + S
Sbjct: 891 NGMLLNDKQVYVGPFLRKQERESTGDRAKFNNVFVKNLSESTTDDELKKTFGEFGT-ITS 1067
Query: 228 IELLKDLQSSGRNRGFAFIEY 248
+++D G+++ F F+ +
Sbjct: 1068AVVMRD--GDGKSKCFGFVNF 1124
Score = 49.3 bits (116), Expect = 2e-06
Identities = 31/153 (20%), Positives = 74/153 (48%), Gaps = 8/153 (5%)
Frame = +3
Query: 118 SEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIE 177
+ +Y+G + NV++ L +G+V VR+ + + GY +V + + A++A++
Sbjct: 444 TSLYVGDLDMNVTDSQLYDLFNQLGQVVSVRVCRDLTTRRSLGYGYVNYSNPQDAARALD 623
Query: 178 ELNNSEFKGKKIKCSSS--------QAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIE 229
LN + + I+ S + +FI N+ K + + + G ++S +
Sbjct: 624 VLNFTPLNNRPIRIMYSHRDPSIRKSGQGNIFIKNLDKAIDHKALHDTFSSFG-NILSCK 800
Query: 230 LLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMS 262
+ D SG+++G+ F+++ A+ + +K++
Sbjct: 801 VAVD--GSGQSKGYGFVQFDTEEAAQKAIEKLN 893
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.303 0.125 0.337
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,174,531
Number of Sequences: 36976
Number of extensions: 92736
Number of successful extensions: 5233
Number of sequences better than 10.0: 550
Number of HSP's better than 10.0 without gapping: 1448
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3207
length of query: 289
length of database: 9,014,727
effective HSP length: 95
effective length of query: 194
effective length of database: 5,502,007
effective search space: 1067389358
effective search space used: 1067389358
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 17 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.8 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0010.10


