

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0008.8
(338 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
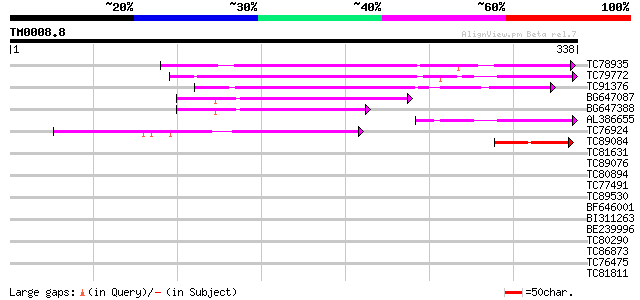
Score E
Sequences producing significant alignments: (bits) Value
TC78935 similar to GP|6760451|gb|AAF28357.1| S-ribonuclease bind... 180 8e-46
TC79772 weakly similar to GP|20259023|gb|AAM14227.1 unknown prot... 133 1e-31
TC91376 weakly similar to GP|17065054|gb|AAL32681.1 putative pro... 114 4e-26
BG647087 weakly similar to GP|22136652|gb unknown protein {Arabi... 89 2e-18
BG647388 weakly similar to GP|22136652|gb| unknown protein {Arab... 71 5e-13
AL386655 weakly similar to GP|20259023|gb| unknown protein {Arab... 67 1e-11
TC76924 similar to GP|21593126|gb|AAM65075.1 inhibitor of apopto... 64 1e-10
TC89084 weakly similar to GP|9759369|dbj|BAB09828.1 gene_id:MYJ2... 46 2e-05
TC81631 similar to GP|20279471|gb|AAM18751.1 unknown protein {Or... 36 0.024
TC89076 similar to GP|18481708|gb|AAL73530.1 hypothetical protei... 35 0.041
TC80894 weakly similar to GP|18855010|gb|AAL79702.1 unknown prot... 35 0.054
TC77491 similar to GP|13937149|gb|AAK50068.1 At1g79380/T8K14_20 ... 33 0.16
TC89530 32 0.27
BF646001 homologue to GP|20303561|gb| putative kinase {Oryza sat... 32 0.35
BI311263 similar to GP|19683021|gb hypothetical protein {Dictyos... 32 0.46
BE239996 similar to GP|21689819|gb| unknown protein {Arabidopsis... 31 0.59
TC80290 weakly similar to GP|13937149|gb|AAK50068.1 At1g79380/T8... 31 0.78
TC86873 homologue to GP|530207|gb|AAA66338.1|| heat shock protei... 30 1.0
TC76475 similar to PIR|T04762|T04762 chitinase homolog T16H5.170... 30 1.3
TC81811 similar to GP|6671365|gb|AAF23176.1| P-glycoprotein {Gos... 30 1.7
>TC78935 similar to GP|6760451|gb|AAF28357.1| S-ribonuclease binding protein
SBP1 {Petunia x hybrida}, partial (93%)
Length = 1304
Score = 180 bits (456), Expect = 8e-46
Identities = 100/251 (39%), Positives = 152/251 (59%), Gaps = 4/251 (1%)
Frame = +1
Query: 91 SASIPNPNALSTGLRLSYDDDERNSSVTSASGSMTAPPSIILSLGDNIRTELDRQQEELD 150
S P ++STGL LS D+ S+ SA S+ +GD+I EL +Q E+D
Sbjct: 382 SVDFLQPRSVSTGLGLSLDNTRLASTGDSALLSL---------IGDDIDRELQQQDLEMD 534
Query: 151 HYIKLQKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNRELAERIKQ 210
++KLQ EQL + + + Q ++ IE +++KLREKE E+EN+N++N EL ++++Q
Sbjct: 535 RFLKLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREKETEVENINKRNMELEDQMEQ 714
Query: 211 VAVEAQNWHYRAKYNESVVNALRNNLQQAISHGVEQGKEGFGDSEVDDAASYIDPN---- 266
++VEA W RA+YNE+++ AL+ NLQQA G KEG GDSEVDD AS +
Sbjct: 715 LSVEAGAWQQRARYNENMIAALKFNLQQAYLQG-RDSKEGCGDSEVDDTASCCNGRSLDF 891
Query: 267 NFLSIPGAPMKSIHPRYQGMENLSCRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQ 326
+ LS + MK + + C+AC+ +V+M+L+PC+HL LCKDC+ ++ CP+CQ
Sbjct: 892 HLLSNENSNMKDL---------MKCKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQ 1044
Query: 327 MIKTASVEVYL 337
K +EVY+
Sbjct: 1045SSKFIGMEVYM 1077
>TC79772 weakly similar to GP|20259023|gb|AAM14227.1 unknown protein
{Arabidopsis thaliana}, partial (33%)
Length = 1480
Score = 133 bits (334), Expect = 1e-31
Identities = 77/256 (30%), Positives = 136/256 (53%), Gaps = 13/256 (5%)
Frame = +2
Query: 96 NPNALSTGLRLSYDDDERNSSVTSASGSMTAPPSIILSLGDNIRTELDRQQEELDHYIKL 155
N N +STGL LS+ D +++ + + L + + +++ +Q++E+D +++
Sbjct: 557 NQNIVSTGLGLSFGD-QQHQRLQLLQQQQCHSSHFLSLLSNGLASQIKQQKDEIDQFLQA 733
Query: 156 QKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNRELAERIKQVAVEA 215
Q E+L R + + +Q++ +++ + E+ + ++LREKE +++ R+N EL R + EA
Sbjct: 734 QGEELQRTIEEKRQRNYRAIIKTAEETVARRLREKEIDLQKATRRNAELEARAAHLRTEA 913
Query: 216 QNWHYRAKYNESVVNALRNNLQQAISHGVEQGKEGFGDSE------------VDDAAS-Y 262
Q W +AK E+ +L+ L A+ G G E G++E +DA S Y
Sbjct: 914 QLWQAKAKEQEATAISLQTQLHHAMMSG---GAENRGENECGLSCALGVEGHAEDAESGY 1084
Query: 263 IDPNNFLSIPGAPMKSIHPRYQGMENLSCRACKAKDVSMLLIPCRHLSLCKDCDGFINVC 322
IDP ++ G+ K CR C + S++++PCRHL +C +CD VC
Sbjct: 1085IDPER--AVVGSGPK-------------CRGCGERVASVVVLPCRHLCVCTECDTRFGVC 1219
Query: 323 PVCQMIKTASVEVYLS 338
PVC +K ++VEVYLS
Sbjct: 1220PVCFTVKNSTVEVYLS 1267
>TC91376 weakly similar to GP|17065054|gb|AAL32681.1 putative protein
{Arabidopsis thaliana}, partial (41%)
Length = 997
Score = 114 bits (286), Expect = 4e-26
Identities = 69/215 (32%), Positives = 108/215 (50%)
Frame = +1
Query: 111 DERNSSVTSASGSMTAPPSIILSLGDNIRTELDRQQEELDHYIKLQKEQLSRGVRDMKQK 170
D S T + +++ PS + N+ L QQ E+D +I E++ + + + K
Sbjct: 358 DSTEESYTQKNIKLSSQPSFV---DQNLLYHLQNQQSEIDLFIAQHTERVRMEIEEQRLK 528
Query: 171 HMASLLTSIEKGIVKKLREKEEEIENMNRKNRELAERIKQVAVEAQNWHYRAKYNESVVN 230
L +I++ + KKL++KEEEI+ M ++N L E+ K + +E Q W A NES VN
Sbjct: 529 QSRMLQAAIQEAVTKKLKQKEEEIQRMEKQNLMLQEKAKTLIMENQIWREMALTNESAVN 708
Query: 231 ALRNNLQQAISHGVEQGKEGFGDSEVDDAASYIDPNNFLSIPGAPMKSIHPRYQGMENLS 290
LRN L+Q ++H VE + DDAAS N+ + ++ P +
Sbjct: 709 TLRNELEQVLAH-VENHRND------DDAASSCGSNHHVKEEVVVEEASSP----VVGKL 855
Query: 291 CRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVC 325
C C ++ +LL+PCRHL LC C I CP+C
Sbjct: 856 CSGCGERESVVLLLPCRHLCLCTMCGTHIRNCPLC 960
>BG647087 weakly similar to GP|22136652|gb unknown protein {Arabidopsis
thaliana}, partial (36%)
Length = 718
Score = 89.0 bits (219), Expect = 2e-18
Identities = 52/149 (34%), Positives = 81/149 (53%), Gaps = 8/149 (5%)
Frame = +1
Query: 100 LSTGLRLSYDDDERNSSVTSAS--------GSMTAPPSIILSLGDNIRTELDRQQEELDH 151
+STGLRLS+DD ++ G ++ S +L G + +++ +Q +ELD
Sbjct: 271 VSTGLRLSFDDQQQQRLQLQLQLQLHQQQQGCHSSSFSSLLPQG--LVSQIKQQHDELDQ 444
Query: 152 YIKLQKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNRELAERIKQV 211
Y++ Q E L R + D +QKH LL + E+ + ++LREKE E R+N EL R Q+
Sbjct: 445 YLQTQGENLRRTLADKRQKHYRELLNAAEEAVARRLREKEVEFAKATRRNAELEARTAQL 624
Query: 212 AVEAQNWHYRAKYNESVVNALRNNLQQAI 240
+EAQ W +A E+ +L+ LQQ I
Sbjct: 625 TMEAQVWQAKAPAQEAAAASLQAQLQQTI 711
>BG647388 weakly similar to GP|22136652|gb| unknown protein {Arabidopsis
thaliana}, partial (26%)
Length = 844
Score = 71.2 bits (173), Expect = 5e-13
Identities = 42/124 (33%), Positives = 67/124 (53%), Gaps = 8/124 (6%)
Frame = +3
Query: 100 LSTGLRLSYDDDERNSSVTSAS--------GSMTAPPSIILSLGDNIRTELDRQQEELDH 151
+STGLRLS+DD ++ G ++ S +L G + +++ +Q +ELD
Sbjct: 432 VSTGLRLSFDDQQQQRLQLQLQLQLHQQQQGCHSSSFSSLLPQG--LVSQIKQQHDELDQ 605
Query: 152 YIKLQKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNRELAERIKQV 211
Y++ Q E L R + D +QKH LL + E+ + ++LREKE E R N EL R Q+
Sbjct: 606 YLQTQGENLRRTLADKRQKHYRELLNAAEEAVARRLREKEVEFAKATRTNAELEARTAQL 785
Query: 212 AVEA 215
+E+
Sbjct: 786 TMES 797
>AL386655 weakly similar to GP|20259023|gb| unknown protein {Arabidopsis
thaliana}, partial (20%)
Length = 498
Score = 66.6 bits (161), Expect = 1e-11
Identities = 34/96 (35%), Positives = 52/96 (53%)
Frame = +1
Query: 243 GVEQGKEGFGDSEVDDAASYIDPNNFLSIPGAPMKSIHPRYQGMENLSCRACKAKDVSML 302
GV G EG + D ++YIDP+ + + A K CR C+ + +++
Sbjct: 46 GVSCGVEGQAE---DAESAYIDPDRVVEVAAARGK-------------CRGCEKRVATVV 177
Query: 303 LIPCRHLSLCKDCDGFINVCPVCQMIKTASVEVYLS 338
++PCRHL +C +CD VCPVC K ++VEV+LS
Sbjct: 178 VLPCRHLCVCTECDAHFRVCPVCFTPKNSTVEVFLS 285
>TC76924 similar to GP|21593126|gb|AAM65075.1 inhibitor of apoptosis-like
protein {Arabidopsis thaliana}, partial (13%)
Length = 759
Score = 63.5 bits (153), Expect = 1e-10
Identities = 51/196 (26%), Positives = 93/196 (47%), Gaps = 11/196 (5%)
Frame = +1
Query: 27 TNQLQLFGNL-QAGRNVDPVSYIGNEHISSMIQPNKRSREMEDISKKQRLQIS--LNYNV 83
TN ++ N+ Q G + P S I +MI P S + + +K + L YNV
Sbjct: 202 TNTNIIYNNISQMGYSSIPPSTIKTTATETMILPPYNSITTDSLPQKTAMNSDSGLTYNV 381
Query: 84 ------CQDDADWSASI--PNPNALSTGLRLSYDDDERNSSVTSASGSMTAPPSIILSLG 135
+D D+S SI P PN+ + ++ R+ + +S S LG
Sbjct: 382 PPLRKRSRDSRDYSNSINFPYPNSYISPSTPQQQNNHRSCASSSFS-----------FLG 528
Query: 136 DNIRTELDRQQEELDHYIKLQKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIE 195
++I ++ RQQ ++D I Q E++ + + +++ L+ +I+ + K+++ EEEIE
Sbjct: 529 EDISLQIQRQQLDIDQLISQQMEKVKYEIEEKRKRQAMRLIQAIDMSVTKRMKG*EEEIE 708
Query: 196 NMNRKNRELAERIKQV 211
+ + N L ER+K +
Sbjct: 709 KIGKMNWALEERVKSL 756
>TC89084 weakly similar to GP|9759369|dbj|BAB09828.1
gene_id:MYJ24.10~ref|NP_055178.1~similar to unknown
protein {Arabidopsis thaliana}, partial (3%)
Length = 1200
Score = 45.8 bits (107), Expect = 2e-05
Identities = 14/47 (29%), Positives = 30/47 (63%)
Frame = +1
Query: 290 SCRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEVY 336
+CR C + +V + ++PC H+ LC+ C ++ CP C++ T ++ ++
Sbjct: 1054 TCRVCLSSEVDITIVPCGHV-LCRKCSSAVSKCPFCRLSSTKAIRIF 1191
>TC81631 similar to GP|20279471|gb|AAM18751.1 unknown protein {Oryza sativa
(japonica cultivar-group)}, partial (40%)
Length = 787
Score = 35.8 bits (81), Expect = 0.024
Identities = 34/140 (24%), Positives = 54/140 (38%), Gaps = 6/140 (4%)
Frame = +1
Query: 193 EIENMNRKNRELAERIKQVAVEAQNWHYRAKYNESV--VNALRNNLQQAISHGVEQGKEG 250
E E ++ N + I Q E + +R K + + VN +R LQ+ G
Sbjct: 229 ETETSSKPNSQ----ITQAVFEKEKGEFRVKVVKQILSVNGMRYELQEIY---------G 369
Query: 251 FGDSEVDDAASYIDPNNFLSIPGAPMKSIHPRYQGMENLSCRACKAKDVSMLLIPCRHLS 310
G+S D +D N QG E C C ++ ++ PCRH+
Sbjct: 370 IGNSVESD----VDDNE----------------QGKE---CVICLSEPRDTIVHPCRHMC 480
Query: 311 LCKDCDGFI----NVCPVCQ 326
+C C + N CP+C+
Sbjct: 481 MCSGCAKVLRFQTNRCPICR 540
>TC89076 similar to GP|18481708|gb|AAL73530.1 hypothetical protein {Sorghum
bicolor}, partial (65%)
Length = 1326
Score = 35.0 bits (79), Expect = 0.041
Identities = 42/190 (22%), Positives = 77/190 (40%), Gaps = 2/190 (1%)
Frame = +2
Query: 149 LDHYIKLQKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNRELAERI 208
+D+ + Q+++ + + D + +A +L + G + + + +++I + + N +
Sbjct: 641 VDNEVSPQEKKTIKAIADASKYPLAIVLVGVGDGPWEDMEKFDDKIPSRDFDNFQFVNFT 820
Query: 209 KQVAVEAQNWHYRAKYNESVVNALRNNLQQAISHGVEQGKEGFGDSEVDDAASYIDPNNF 268
K + ++N K VNAL + VE K G V A I P
Sbjct: 821 K---IMSKNTSAAEKEAAFAVNALME-IPFQYKACVELQKLG----RVTGRAKRIVPKPS 976
Query: 269 LSIPGAPMKSIHPRYQGMENLSCRAC--KAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQ 326
P S ++ C C AKD L C H++ C+DC + CP+C+
Sbjct: 977 PVPYSRPAHSNSTTDDQNQSACCPVCLTNAKD---LAFGCGHMT-CRDCGSRLRHCPICR 1144
Query: 327 MIKTASVEVY 336
T+ + VY
Sbjct: 1145HRITSRLRVY 1174
>TC80894 weakly similar to GP|18855010|gb|AAL79702.1 unknown protein {Oryza
sativa}, partial (20%)
Length = 679
Score = 34.7 bits (78), Expect = 0.054
Identities = 33/141 (23%), Positives = 59/141 (41%), Gaps = 5/141 (3%)
Frame = +3
Query: 104 LRLSYDDDERNS-SVTSASGSMTAPPSIILSLGDNIRTELDRQQEELDHYIKLQKEQLSR 162
LR D +ER +T A + A +S + I + Q + ++ E+
Sbjct: 183 LRAQLDANERKKLEMTEAIKAKEAEAETYISEIETIGQAYEDMQTQHQRLLQQVAERDDC 362
Query: 163 GVR----DMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNRELAERIKQVAVEAQNW 218
++ MK KH+ S L S ++ V +L++ IEN ++ E+IK + EA
Sbjct: 363 NIKLVSESMKAKHLHSTLLSEKQAFVDQLQKINSLIENSKKRIANSEEQIKHILSEAAKC 542
Query: 219 HYRAKYNESVVNALRNNLQQA 239
+ K+ + + R L A
Sbjct: 543 THDEKHLAAALKFARWELADA 605
>TC77491 similar to GP|13937149|gb|AAK50068.1 At1g79380/T8K14_20 {Arabidopsis
thaliana}, partial (81%)
Length = 1537
Score = 33.1 bits (74), Expect = 0.16
Identities = 23/75 (30%), Positives = 37/75 (48%)
Frame = +2
Query: 262 YIDPNNFLSIPGAPMKSIHPRYQGMENLSCRACKAKDVSMLLIPCRHLSLCKDCDGFINV 321
++ P +FLS P S+ + ME C K KD L C H++ C+DC ++
Sbjct: 1055 HMPPEHFLS--RMPTSSMDDQRNQMECAICLTNK-KD---LAFGCGHMT-CRDCGSRLSD 1213
Query: 322 CPVCQMIKTASVEVY 336
CP+C+ T + V+
Sbjct: 1214 CPICRQRITNRLRVF 1258
>TC89530
Length = 1614
Score = 32.3 bits (72), Expect = 0.27
Identities = 26/94 (27%), Positives = 44/94 (46%), Gaps = 3/94 (3%)
Frame = +2
Query: 110 DDERNSSVTSASGSMTAPPSIILS--LGDNIRTELDRQQEELDHYIKLQKEQLSRGVRDM 167
D ++ TS+SG + I D + EL R + E + KL E+ S + +
Sbjct: 476 DTDKRKKGTSSSGKKKSSSKIAEKEKCPDETKDELSRVKAECE---KLVAEKDSELLALL 646
Query: 168 KQKHMA-SLLTSIEKGIVKKLREKEEEIENMNRK 200
++K+ + +E KKLR K++E+E N K
Sbjct: 647 QEKNFVWNQFNKMETDYTKKLRSKQKEVEKTNEK 748
>BF646001 homologue to GP|20303561|gb| putative kinase {Oryza sativa
(japonica cultivar-group)}, partial (6%)
Length = 667
Score = 32.0 bits (71), Expect = 0.35
Identities = 21/74 (28%), Positives = 38/74 (50%)
Frame = +2
Query: 145 QQEELDHYIKLQKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNREL 204
+QE ++ +K+++ + GV +MK+ S IE+ + +K+ EKE E+E EL
Sbjct: 404 KQENVEMEMKVRELERRIGVIEMKEVEENSKRVRIEEEMKEKIYEKEMEVE-------EL 562
Query: 205 AERIKQVAVEAQNW 218
+ VE + W
Sbjct: 563 KSMLMGKKVEEEKW 604
>BI311263 similar to GP|19683021|gb hypothetical protein {Dictyostelium
discoideum}, partial (1%)
Length = 730
Score = 31.6 bits (70), Expect = 0.46
Identities = 20/96 (20%), Positives = 47/96 (48%)
Frame = +3
Query: 143 DRQQEELDHYIKLQKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNR 202
++Q +E D I +Q Q+ ++ ++ + L ++E + + + EEI + +KN+
Sbjct: 318 EKQGQETDQLINIQNGQIKLMLQHHIRELQVATLKNMEIYSRQIMTKANEEIAKVVKKNQ 497
Query: 203 ELAERIKQVAVEAQNWHYRAKYNESVVNALRNNLQQ 238
E+ ++ + E + A+ + AL N L++
Sbjct: 498 EMENLLRSLETEKRFLKRIAEERGATTIALHNKLEE 605
>BE239996 similar to GP|21689819|gb| unknown protein {Arabidopsis thaliana},
partial (16%)
Length = 420
Score = 31.2 bits (69), Expect = 0.59
Identities = 19/72 (26%), Positives = 30/72 (41%), Gaps = 1/72 (1%)
Frame = +2
Query: 266 NNFLSIPGAP-MKSIHPRYQGMENLSCRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPV 324
N F++ P SI P C C M C H + CK+C +++CP+
Sbjct: 146 NAFIATPRITNFDSIEPTAPASAEPVCPICLTNPKDMAF-GCGHTT-CKECGATLSLCPM 319
Query: 325 CQMIKTASVEVY 336
C+ T + +Y
Sbjct: 320 CRQQITTRLRLY 355
>TC80290 weakly similar to GP|13937149|gb|AAK50068.1 At1g79380/T8K14_20
{Arabidopsis thaliana}, partial (10%)
Length = 1735
Score = 30.8 bits (68), Expect = 0.78
Identities = 37/190 (19%), Positives = 75/190 (39%), Gaps = 6/190 (3%)
Frame = +1
Query: 153 IKLQKEQLSRGVRDMKQ------KHMASLLTSIEKGIVKKLREKEEEIENMNRKNRELAE 206
+K + E +S + +Q + + L ++E G+ + R+K+EE +M +
Sbjct: 991 LKHENELISNAEKQARQMVTNLSQRLHCLQIAMETGVAE--RKKQEEFIHMLQN------ 1146
Query: 207 RIKQVAVEAQNWHYRAKYNESVVNALRNNLQQAISHGVEQGKEGFGDSEVDDAASYIDPN 266
+ A + +H + KY V LR L + +E + A D +
Sbjct: 1147 ---ECAKTKREFHEQNKY----VERLRQKLDNVPQFPIRMAEETKQKLTIPSKAILADES 1305
Query: 267 NFLSIPGAPMKSIHPRYQGMENLSCRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQ 326
+ S+ R + + SC C + + L C H++ C++C I C +C+
Sbjct: 1306 KAAVVEVYKTISLS-RNPTLTSQSCTICLSNEKD-LAFGCGHMT-CRECGSKIRKCHICR 1476
Query: 327 MIKTASVEVY 336
T + ++
Sbjct: 1477 KKITDRIRLF 1506
>TC86873 homologue to GP|530207|gb|AAA66338.1|| heat shock protein {Glycine
max}, partial (53%)
Length = 1475
Score = 30.4 bits (67), Expect = 1.0
Identities = 32/119 (26%), Positives = 55/119 (45%), Gaps = 21/119 (17%)
Frame = +2
Query: 137 NIRTELDRQQEELD-------------HYIKLQKEQLSRGVRDMKQKHMASLLTSIEKGI 183
N+R +LD Q EE+D H ++ +K++ S+ ++ + L ++
Sbjct: 95 NVRVQLDSQPEEIDNLERKRMQLEVELHALEKEKDKASKARLVDVRRELDDLRDKLQPLK 274
Query: 184 VKKLREKE--EEIENMNRKNREL------AERIKQVAVEAQNWHYRAKYNESVVNALRN 234
+K +EKE +EI + +K EL AER +A A + Y A E V A++N
Sbjct: 275 MKYSKEKERIDEIRRLKQKREELLFALQEAERRYDLA-RAADLRYGA--IEEVETAIKN 442
>TC76475 similar to PIR|T04762|T04762 chitinase homolog T16H5.170 -
Arabidopsis thaliana, partial (51%)
Length = 1496
Score = 30.0 bits (66), Expect = 1.3
Identities = 25/81 (30%), Positives = 38/81 (46%)
Frame = +3
Query: 188 REKEEEIENMNRKNRELAERIKQVAVEAQNWHYRAKYNESVVNALRNNLQQAISHGVEQG 247
RE+E E E + RE ER + + Y K++ +++ L LQ +S
Sbjct: 93 RERERERERERERERE-RERPRAEFGTRKKMAYSKKHSFLLISTLLMILQLQLSFTNAMI 269
Query: 248 KEGFGDSEVDDAASYIDPNNF 268
K G+ S+ AAS IDP+ F
Sbjct: 270 KGGYWYSDSGLAASDIDPSYF 332
>TC81811 similar to GP|6671365|gb|AAF23176.1| P-glycoprotein {Gossypium
hirsutum}, partial (17%)
Length = 850
Score = 29.6 bits (65), Expect = 1.7
Identities = 14/32 (43%), Positives = 20/32 (61%)
Frame = +1
Query: 239 AISHGVEQGKEGFGDSEVDDAASYIDPNNFLS 270
+I + GKEG DSEV +AA + +NF+S
Sbjct: 199 SIYENILYGKEGASDSEVIEAAKLANAHNFIS 294
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.131 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,705,241
Number of Sequences: 36976
Number of extensions: 127470
Number of successful extensions: 735
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 722
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 726
length of query: 338
length of database: 9,014,727
effective HSP length: 97
effective length of query: 241
effective length of database: 5,428,055
effective search space: 1308161255
effective search space used: 1308161255
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0008.8


