

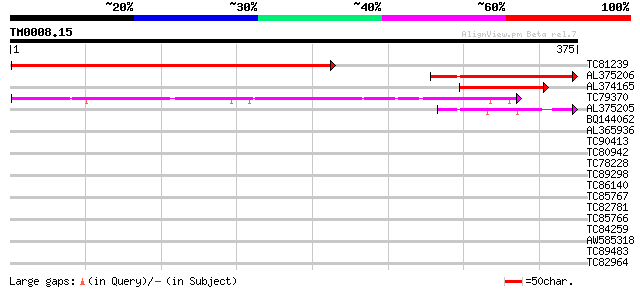
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0008.15
(375 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC81239 weakly similar to PIR|G86367|G86367 protein F28C11.3 [im... 300 5e-82
AL375206 135 2e-32
AL374165 97 1e-20
TC79370 87 1e-17
AL375205 48 7e-06
BQ144062 weakly similar to SP|P03211|EBN1_ EBNA-1 nuclear protei... 34 0.10
AL365936 30 1.5
TC90413 similar to GP|21593314|gb|AAM65263.1 unknown {Arabidopsi... 30 1.5
TC80942 GP|21112489|gb|AAM40721.1 conserved hypothetical protein... 29 2.6
TC78228 similar to GP|10177770|dbj|BAB11102. protein kinase-like... 29 2.6
TC89298 similar to PIR|B96698|B96698 unknown protein F12B7.3 [im... 29 3.4
TC86140 similar to GP|9757759|dbj|BAB08240.1 emb|CAB55405.1~gene... 28 4.4
TC85767 28 5.7
TC82781 similar to PIR|F96767|F96767 proteinase IV F2P9.14 [impo... 28 5.7
TC85766 28 5.7
TC84259 weakly similar to PIR|T43257|T43257 beta-1 3 exoglucanas... 28 7.5
AW585318 similar to PIR|T04590|T045 hypothetical protein F23E13.... 28 7.5
TC89483 similar to GP|18377662|gb|AAL66981.1 unknown protein {Ar... 27 9.8
TC82964 similar to GP|22655058|gb|AAM98120.1 predicted protein {... 27 9.8
>TC81239 weakly similar to PIR|G86367|G86367 protein F28C11.3 [imported] -
Arabidopsis thaliana, partial (18%)
Length = 1002
Score = 300 bits (769), Expect = 5e-82
Identities = 146/214 (68%), Positives = 168/214 (78%)
Frame = +3
Query: 2 AAQWELPEEAPIHGDVLETILSHVPLIHLVPACHVSNTWKHAVFSSLAHTRTIKPWLIIL 61
A + E +EAPIHGDVLETI S VPLI LVP+CHVS +W VFSSL H R IKPWLI+L
Sbjct: 360 AIKREPKQEAPIHGDVLETIFSFVPLIDLVPSCHVSKSWNKTVFSSLTHVRQIKPWLIVL 539
Query: 62 TQSTRASHVITAHAYDPRSHVWVEMKLPHPLKGHPSATAAVRSSHSTLLYTLSPAEFAFS 121
Q++RAS V AHAYDPRSH W+++ PL A+RSSHSTLLYTLSP+EF FS
Sbjct: 540 PQTSRASRVTIAHAYDPRSHAWLQITKHQPLINKTQEIPAIRSSHSTLLYTLSPSEFTFS 719
Query: 122 IDALHLDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVMCQ 181
+DAL L+WH AP+PRVWRTDPIVARVG VVVAGGAC+FEDDPLAVEMY+MES WV CQ
Sbjct: 720 LDALQLEWHKAPSPRVWRTDPIVARVGNCVVVAGGACEFEDDPLAVEMYNMESRDWVNCQ 899
Query: 182 SMPAMLKGSSGSTWLSVAVAGEEVLVTEESSGMT 215
SMP LK SS S+WLSVAV GE +LVTE++S +T
Sbjct: 900 SMPMTLKTSSASSWLSVAVVGETMLVTEKNSRVT 1001
>AL375206
Length = 451
Score = 135 bits (341), Expect = 2e-32
Identities = 64/97 (65%), Positives = 80/97 (81%)
Frame = +2
Query: 279 EEVGEMPEELLEKFKGDSDELGSLEVTWVGNFVYLRNTLEIEELVMCEVVNGSRCEWRSV 338
EE+G MP+E++EK +GDS E GS+E WVGN VYLRNTL ++ELV+CEVVNG+ CEWRSV
Sbjct: 8 EEIGSMPKEMVEKLRGDS-EFGSVEAIWVGNLVYLRNTLVLDELVVCEVVNGNLCEWRSV 184
Query: 339 RNAAVDGGTRMVVCGSEVRLEDLQSAMVSGIQTCRTK 375
+NAAVDGGTRMV CG +V++EDLQ A++S QT K
Sbjct: 185 KNAAVDGGTRMVFCGGDVKMEDLQRAVLSEKQTFSMK 295
>AL374165
Length = 180
Score = 97.1 bits (240), Expect = 1e-20
Identities = 44/59 (74%), Positives = 50/59 (84%)
Frame = +2
Query: 298 ELGSLEVTWVGNFVYLRNTLEIEELVMCEVVNGSRCEWRSVRNAAVDGGTRMVVCGSEV 356
E GS+EV WVG+FVYLRNTL +EELV+CEV+NGS CEWRSVRN AVDGG MV CG +V
Sbjct: 2 EFGSVEVIWVGDFVYLRNTLVLEELVVCEVMNGSLCEWRSVRNVAVDGGPGMVFCGGDV 178
>TC79370
Length = 1557
Score = 87.0 bits (214), Expect = 1e-17
Identities = 90/360 (25%), Positives = 155/360 (43%), Gaps = 23/360 (6%)
Frame = +1
Query: 2 AAQWELPEEAPIHGDVLETILSHVPLIHLVPACHVSNTWKHAVFSSLA----HTRTIKPW 57
++Q + E + D++E+ILSH+P+ L+ A V W + + SS + H + KPW
Sbjct: 106 SSQTNMTEITNLSLDLIESILSHLPIPSLIQASTVCKLW-YTILSSSSFSSNHNQKHKPW 282
Query: 58 LIILTQSTRASHVITAHAYDPRSHVWVEMKLPHPLKGHPSATAAVRSSHSTLLYTLSPAE 117
+ +S + A+DP S+ W + P +P+ T+ + +S + ++
Sbjct: 283 FFLHGIHNISSKNNQSFAFDPSSNSWFLLPTPQQPLHYPNNTSFIGTSS---YFFITAPN 453
Query: 118 FAFSIDALHLDWHHAPAPRVWRTDPIVA----RVGQRVVVAGGA------CDFEDDPLAV 167
F ++ L W P R +P++ + + +V GG D E D L V
Sbjct: 454 FVYTSILRPLAWSSTPPLHFPRINPLLGVFNDGLSLKFIVVGGVRFIGNLVDIE-DRLDV 630
Query: 168 EMYDMESNTWVMCQSMPAMLKGSSGSTWLSVAVAGEEVLVTEESSGMTFCFDTVSMKWDG 227
E+YD +W + +P + + S+ LS A+ + V S FD W
Sbjct: 631 EIYDPLLGSWDLAPPLPVDFRSGNSSSSLSSALFKGKFFVFGIYSCFVSSFDLKLRVWSD 810
Query: 228 PYDLCPDQSVGYCVTGMLGKRLMVAGVVGDGEDVKAVKLWAVKGGLGSGMTEEVGEMPEE 287
+ P V + +RL++AGV + + LW V S E+G MP +
Sbjct: 811 VRIVRP-SGVVFSFLIACRERLVLAGVC-NSPSGSSFNLWEVDE--KSMEICEIGVMPHD 978
Query: 288 LLEK-FKGDSDE-LGSLEVTWVGNFVYLRNT--LEIEELVMCEVVN-----GSRCEWRSV 338
LL F GD D+ SL+ +G+ +Y+ N + +CE+ + S+C WR V
Sbjct: 979 LLSSLFDGDEDDRFASLKCVGLGDLIYVFNEDYHRMYPACVCEIRSRGGGENSKCYWRRV 1158
>AL375205
Length = 508
Score = 47.8 bits (112), Expect = 7e-06
Identities = 39/103 (37%), Positives = 53/103 (50%), Gaps = 11/103 (10%)
Frame = +2
Query: 284 MPEELLEKFKGDSDELGSLEVTWVGNFVYLR------NTLEIE-ELVMCEVVNGSRCE-- 334
MP+E++EK +GDS E GS+E WVGN VYLR + L+ +V E+V +CE
Sbjct: 23 MPKEMVEKLRGDS-EFGSVEAIWVGNLVYLR*HDWCWSELDCHVTVVKWELV*VEKCEEC 199
Query: 335 --WRSVRNAAVDGGTRMVVCGSEVRLEDLQSAMVSGIQTCRTK 375
R + GG + C EDLQ A++S QT K
Sbjct: 200 GGGRWNEDGVYVGG*MLTNC------EDLQRAVLSEKQTFSMK 310
>BQ144062 weakly similar to SP|P03211|EBN1_ EBNA-1 nuclear protein. [strain
B95-8 Human herpesvirus 4] {Epstein-barr virus},
partial (8%)
Length = 1390
Score = 33.9 bits (76), Expect = 0.10
Identities = 21/63 (33%), Positives = 30/63 (47%), Gaps = 7/63 (11%)
Frame = -2
Query: 60 ILTQSTRASHVITAHAYDPRSHVWVEMKL------PHPLKGHPSATAAVR-SSHSTLLYT 112
I+T T ++H+ +H+ PR H + L P P +GH ATA R H+ L
Sbjct: 837 IVTPRTTSTHLSASHSATPRLHDHIASLLRALARAPRPARGHAGATAPARPRPHAPLAAP 658
Query: 113 LSP 115
L P
Sbjct: 657 LRP 649
>AL365936
Length = 441
Score = 30.0 bits (66), Expect = 1.5
Identities = 11/41 (26%), Positives = 23/41 (55%)
Frame = +1
Query: 97 SATAAVRSSHSTLLYTLSPAEFAFSIDALHLDWHHAPAPRV 137
++T + S S +YT +P + + + D+ +APAP++
Sbjct: 253 ASTPKIEKSKSEAVYTPNPTKSDYEVPKTKTDYEYAPAPKI 375
>TC90413 similar to GP|21593314|gb|AAM65263.1 unknown {Arabidopsis
thaliana}, partial (64%)
Length = 1104
Score = 30.0 bits (66), Expect = 1.5
Identities = 32/142 (22%), Positives = 54/142 (37%), Gaps = 11/142 (7%)
Frame = +1
Query: 95 HPSATAAVRSSHSTLLYTLSPAEF--AFSIDALHLDWHHAPAPRVW--RTDPIVAR---- 146
+P + + S+ +L L P F+ D + W + P + W R +V R
Sbjct: 571 YPQLESCLSSAVEVMLLILRPVITMGCFATDEV---WSYDPIIQQWAPRASMLVPRSMFA 741
Query: 147 ---VGQRVVVAGGACDFEDDPLAVEMYDMESNTWVMCQSMPAMLKGSSGSTWLSVAVAGE 203
+ ++VVAGG EMYD E + W MP + + + + S V G
Sbjct: 742 CCVLNGKIVVAGGFTSCRKTISQAEMYDPEKDVWT---PMPDLHRTQNSA--CSGIVIGG 906
Query: 204 EVLVTEESSGMTFCFDTVSMKW 225
++ V + D +W
Sbjct: 907 KMHVLHKDMSTVQVLDNAGARW 972
>TC80942 GP|21112489|gb|AAM40721.1 conserved hypothetical protein
{Xanthomonas campestris pv. campestris str. ATCC 33913},
partial (9%)
Length = 482
Score = 29.3 bits (64), Expect = 2.6
Identities = 11/39 (28%), Positives = 21/39 (53%)
Frame = -2
Query: 99 TAAVRSSHSTLLYTLSPAEFAFSIDALHLDWHHAPAPRV 137
T + S S +YT +P + + + D+ +APAP++
Sbjct: 208 TPKIEKSKSEAVYTPNPTKSDYEVPKTKTDYEYAPAPKI 92
>TC78228 similar to GP|10177770|dbj|BAB11102. protein kinase-like {Arabidopsis
thaliana}, partial (90%)
Length = 2066
Score = 29.3 bits (64), Expect = 2.6
Identities = 20/54 (37%), Positives = 26/54 (48%), Gaps = 5/54 (9%)
Frame = -3
Query: 96 PSATAAVRSSHSTLLYTLSPAEFAFSIDALHLDWHH-----APAPRVWRTDPIV 144
PS S S+LL+ S F + A L HH +PAP VW++DP V
Sbjct: 1455 PSPEKQNHPSSSSLLFAASGWS-TFGMKAQ*LPPHHLYTQFSPAPHVWKSDPCV 1297
>TC89298 similar to PIR|B96698|B96698 unknown protein F12B7.3 [imported] -
Arabidopsis thaliana, partial (31%)
Length = 898
Score = 28.9 bits (63), Expect = 3.4
Identities = 16/47 (34%), Positives = 25/47 (53%)
Frame = +1
Query: 137 VWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVMCQSM 183
V R D A V V + GG D+ +VEMYD +++ W + +S+
Sbjct: 19 VARYDFACAEVDGLVYIVGGYGVNGDNLSSVEMYDPDTDKWTLIESL 159
>TC86140 similar to GP|9757759|dbj|BAB08240.1
emb|CAB55405.1~gene_id:MUF9.20~similar to unknown protein
{Arabidopsis thaliana}, partial (88%)
Length = 1825
Score = 28.5 bits (62), Expect = 4.4
Identities = 16/48 (33%), Positives = 24/48 (49%), Gaps = 1/48 (2%)
Frame = +3
Query: 167 VEMYDMESNTWVMCQSMPAMLKGSSGSTW-LSVAVAGEEVLVTEESSG 213
V+MYD + NTW +P S+G W L+ G+++LV G
Sbjct: 1095 VKMYDKQKNTWNELGRLPVRADSSNG--WGLAFKACGDKLLVVGGQRG 1232
>TC85767
Length = 717
Score = 28.1 bits (61), Expect = 5.7
Identities = 11/34 (32%), Positives = 17/34 (49%)
Frame = -3
Query: 26 PLIHLVPACHVSNTWKHAVFSSLAHTRTIKPWLI 59
PL+ +V +NTW + + + R I PW I
Sbjct: 481 PLVSIVQLIQTTNTWPTSTYKTRHMKRFIHPWTI 380
>TC82781 similar to PIR|F96767|F96767 proteinase IV F2P9.14 [imported] -
Arabidopsis thaliana, partial (19%)
Length = 692
Score = 28.1 bits (61), Expect = 5.7
Identities = 14/50 (28%), Positives = 24/50 (48%)
Frame = -1
Query: 12 PIHGDVLETILSHVPLIHLVPACHVSNTWKHAVFSSLAHTRTIKPWLIIL 61
P HG +L S L+ +P+C S WK + + + +++ W I L
Sbjct: 173 PNHG-MLHPYRSKPSLVQPLPSCLQSLIWKKPPYHEIVYKNSVQTWQIAL 27
>TC85766
Length = 1494
Score = 28.1 bits (61), Expect = 5.7
Identities = 11/34 (32%), Positives = 17/34 (49%)
Frame = -3
Query: 26 PLIHLVPACHVSNTWKHAVFSSLAHTRTIKPWLI 59
PL+ +V +NTW + + + R I PW I
Sbjct: 1021 PLVSIVQLIQTTNTWPTSTYKTRHMKRFIHPWTI 920
>TC84259 weakly similar to PIR|T43257|T43257 beta-1 3 exoglucanase (EC
3.2.1.-) precursor - fungus (Trichoderma harzianum),
partial (8%)
Length = 788
Score = 27.7 bits (60), Expect = 7.5
Identities = 20/60 (33%), Positives = 30/60 (49%), Gaps = 1/60 (1%)
Frame = -1
Query: 20 TILSHVPLIHLVPACHVSNTWKHAVFSSLAHTRTIKPWLIIL-TQSTRASHVITAHAYDP 78
+++S + HL PA +W+HAV SL + P+ STR+ H I +H DP
Sbjct: 599 SLMSRILC*HLAPA----PSWQHAVL*SLKKEYSPAPYATTFRLPSTRSPHAI-SHGGDP 435
>AW585318 similar to PIR|T04590|T045 hypothetical protein F23E13.100 -
Arabidopsis thaliana, partial (19%)
Length = 585
Score = 27.7 bits (60), Expect = 7.5
Identities = 9/22 (40%), Positives = 15/22 (67%)
Frame = +3
Query: 40 WKHAVFSSLAHTRTIKPWLIIL 61
W+H + SS R ++PWL++L
Sbjct: 468 WQHGLMSSGQKWRPLRPWLLVL 533
>TC89483 similar to GP|18377662|gb|AAL66981.1 unknown protein {Arabidopsis
thaliana}, partial (35%)
Length = 1711
Score = 27.3 bits (59), Expect = 9.8
Identities = 14/46 (30%), Positives = 22/46 (47%)
Frame = -3
Query: 20 TILSHVPLIHLVPACHVSNTWKHAVFSSLAHTRTIKPWLIILTQST 65
++ S V + ++ AC SL+H RT PW ++ QST
Sbjct: 1346 SLKSTVSFVDVIDACRT--------LPSLSHMRTTPPWSLVSYQST 1233
>TC82964 similar to GP|22655058|gb|AAM98120.1 predicted protein {Arabidopsis
thaliana}, partial (73%)
Length = 1244
Score = 27.3 bits (59), Expect = 9.8
Identities = 12/41 (29%), Positives = 24/41 (58%), Gaps = 1/41 (2%)
Frame = +3
Query: 149 QRVVVAGGACDFEDDPL-AVEMYDMESNTWVMCQSMPAMLK 188
+ + + G CD + L + E+Y+ E+ TW +++P M+K
Sbjct: 573 REIAILAGGCDSDGHILDSAELYNSENQTW---ETLPNMIK 686
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.133 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,191,259
Number of Sequences: 36976
Number of extensions: 194111
Number of successful extensions: 974
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 962
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 971
length of query: 375
length of database: 9,014,727
effective HSP length: 98
effective length of query: 277
effective length of database: 5,391,079
effective search space: 1493328883
effective search space used: 1493328883
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0008.15


